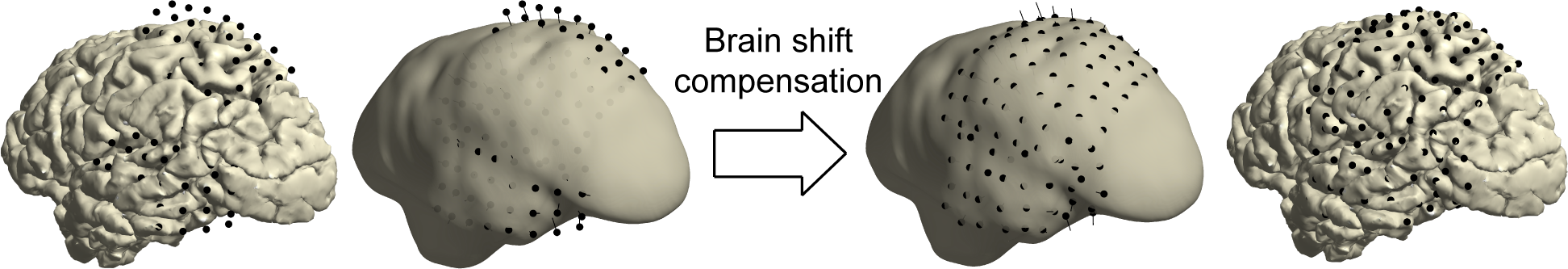Analysis of human ECoG and sEEG recordings
Introduction
Intracranial EEG (iEEG) allows simultaneous recordings from tens to hundreds of electrodes placed directly on the neocortex (electrocorticography, ECoG), or intracortically (stereoelectroencephalography, SEEG). These recordings are known for known for their high spatiotemporal precision. In humans, the most common implementation of iEEG is when non-invasive techniques such as scalp-EEG and MRI do not provide sufficient information to guide surgery in medication refractory epilepsy patients. This tutorial illustrates how to deal with the multitude of raw anatomical and electrophysiological data files in order to get to integrated neural observations.
Before we start, it is important to emphasize that human iEEG datasets are solely acquired for clinical purposes and come in different shapes and sizes. Some medical institutes use photography or X-ray (e.g., see Analysis of monkey ECoG recordings) for including anatomy in the analysis of the functional recordings, whilst others use CT (3D image from a series of X-rays) and/or MR, or combinations thereof. The example iEEG dataset used in this tutorial is not representative for all the datasets obtained in the field but it is meant to serve as a platform for thinking and dealing with the challenges associated with analyzing this type of data.
The tutorial demonstrates the analysis of task-related high-frequency-band activity (~70 to 150 Hz), a prominent neural signature in intracranial data that has been associated with neuron population level firing rate. Many other supported analyses such as event-related potential analysis, connectivity analysis, and statistical analysis have been described in detail elsewhere (Oostenveld et al., 2011; Maris & Oostenveld, 2007; Bastos & Schoffelen, 2016). You will need the iEEG data of SubjectUCI29, which can be obtained from here. If you are getting started with FieldTrip, download the most recent version from its homepage or GitHub and set up your MATLAB path.
The information on this page originates from the human intracranial data analysis protocol described in Stolk, Griffin et al., Integrated analysis of anatomical and electrophysiological human intracranial data, Nature Protocols, 2018. Please cite that paper when you use the methods described here.
Background
The example iEEG data set was acquired at the Medical Center of the University of California, Irvine. The Office for the Protection of Human Subjects of the University of California, Berkeley, approved the study and the subject gave informed consent. The data set includes a pre-implant T1-weighted MRI, a post-implant CT, a post-implant T1-weighted MRI, and neural recordings from 96 ECoG and 56 SEEG electrodes that were implanted as part of the preparation for the epilepsy surgery. The neural data were recorded in the context of an experiment that required the patient to press a button with the right hand when hearing a target tone.
This particular iEEG dataset was chosen for three reasons. First, it contains neural recordings from both cortical grid (ECoG) and stereotactically inserted depth electrodes (SEEG), requiring strategies for dealing with each type as well as their combination in the analysis. Second, the pre-implant MRI is not of the best quality (a contrast agent was used), electrodes of adjacent cortical grids have seemingly merged with one another in the post-implant CT, and there is significant electrode displacement due to a subdural hygroma contributing to so-called “brain shift”. These issues reflect real-world challenges in intracranial data analysis, allowing demonstrating the analysis of non-ideal data. Finally, the experimental paradigm is simple enough to need no further explanation, yet requires performing all the fundamental steps underlying the analysis of intracranial data recorded using a more complex experimental paradigm.
The dataset used in this tutorial
The original dataset (after defacing the imaging data with ft_defacevolume) and the processed results are available for download from zenodo. Raw DICOM images and recording files are not shared to protect the subject’s identity.
The individual files can also be downloaded from our download server.
Procedure
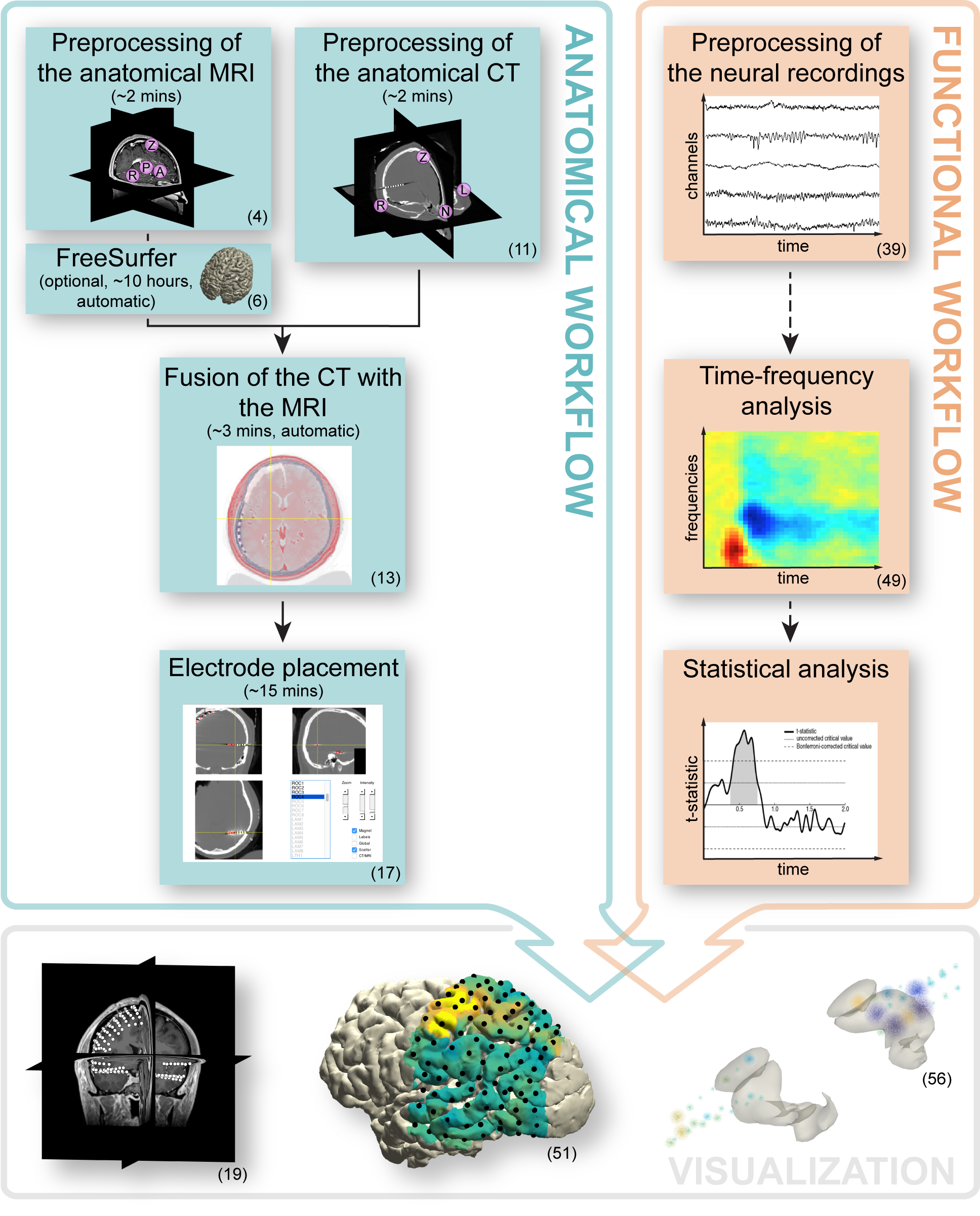
The tutorial has two parallel but interrelated workflows, as shown in the figure below. The first workflow entails the processing of anatomical data. Its main activities constitute the preprocessing and fusion of the anatomical images, and electrode placement (Steps 1-20). Secondary activities that are also demonstrated include cortical surface extraction with FreeSurfer, brain shift compensation, spatial normalization, and anatomical labeling (Steps 6 and 21-34). Generally, the anatomical workflow aims to obtain estimates of the electrode locations in relation to the individual and atlas-based brain anatomy, which is a one-time procedure for each subject.
The second workflow focuses on improving the signal-to-noise ratio and extracting the relevant features from the electrophysiological data, while preparing for subsequent analyses. It minimally encompasses the preprocessing of the neural recordings, but may also include follow-up activities such as time-frequency and single-subject or group-level statistical analysis (Steps 35-46). Generally, the specifics of the functional workflow depend ultimately on the clinical or research question at hand and contingencies in the experimental paradigm.
The two workflows become intrinsically connected for the first time during the electrode placement activity (Step 17), which offers the opportunity to directly link anatomical locations to electrode labels corresponding to the neural recordings. This activity involves a graphical user interface designed for efficient yet precise identification of electrodes in even the most challenging cases. The integration of the two workflows culminates in an interactive and anatomically informed data exploration tool and the ability to represent functional and epileptiform neural activity overlaid on cortical and subcortical surface models, in figure or video format (Steps 47-57).
Anatomical workflow
1) Specify the subject ID. This ID will be used in the file naming, in addition to information about the type of data (e.g., MRI, CT), the coordinate system it is in (e.g., ACPC, MNI), and the process(es) that were applied to it (e.g., f for fusion). For example, a CT scan that is aligned to the ACPC coordinate system and that has just been fused with the anatomical MRI is written out to file as subjID_CT_acpc_f.nii.
subjID = 'SubjectUCI29';
Preprocessing of the anatomical MRI
2) Import the anatomical MRI into the MATLAB workspace using ft_read_mri. The MRI comes in the format of a single file with an .img or .nii extension, or a folder containing a series of files with a .dcm or .ima extension (DICOM; Supplementary File 2 of the original paper may aid in the search and visualization of a DICOM series). For tutorial purposes, we read in the already preprocessed MRI:
mri = ft_read_mri([subjID '_MR_acpc.nii']); % we used the dcm series
3) Determine the native orientation of the anatomical MRI’s left-right axis using ft_determine_coordsys (Box 3 and Supplementary Video 1 of the original paper).
mri = ft_determine_coordsys(mri);
CRITICAL STEP To correctly fuse the MRI and CT scans at a later step, accuracy in demarcating the right hemisphere landmark in the following step is important for avoiding an otherwise hard to detect flip of the scan’s left and right orientation.
4) Align the anatomical MRI to the ACPC coordinate system, a preferred convention for the FreeSurfer operation optionally used in a later step. In this coordinate system, the origin (coordinate [0,0,0]) is at the anterior commissure (AC), the Y-axis runs along the line between the anterior and posterior commissure (PC), and the Z-axis lies in the midline dividing the two cerebral hemispheres. Specify the anterior and posterior commissure, an interhemispheric location along the midline at the top of the brain, and a location in the brain’s right hemisphere.
If the scan was found to have a left-to-right orientation in the previous step, the right hemisphere is identified as the hemisphere having larger values along the left-right axis. Vice versa, in a right-to-left system, the right hemisphere has smaller values along that axis than its left counterpart (Supplementary Video 2).
cfg = [];
cfg.method = 'interactive';
cfg.coordsys = 'acpc';
mri_acpc = ft_volumerealign(cfg, mri);
5) Write the preprocessed anatomical MRI out to file.
cfg = [];
cfg.filename = [subjID '_MR_acpc'];
cfg.filetype = 'nifti';
cfg.parameter = 'anatomy';
ft_volumewrite(cfg, mri_acpc);
Cortical surface extraction with FreeSurfer (optional)
6) Execute FreeSurfer’s recon-all functionality from the Linux or MacOS terminal (Windows via VirtualBox), or from the MATLAB command window as below. This set of commands will create a folder named ‘freesurfer’ in the subject directory, with subdirectories containing a multitude of FreeSurfer-generated files.
For tutorial purposes, the example dataset contains the output from FreeSurfer, a folder named ‘freesurfer’, for continuation with the protocol. You can therefore skip this time-consuming computation and continue with step 7.
fshome = <path to freesurfer home directory>;
subdir = <path to subject directory>;
mrfile = <path to subject MR_acpc.nii>;
system(['export FREESURFER_HOME=' fshome '; ' ...
'source $FREESURFER_HOME/SetUpFreeSurfer.sh; ' ...
'mri_convert -c -oc 0 0 0 ' mrfile ' ' [subdir '/tmp.nii'] '; ' ...
'recon-all -i ' [subdir '/tmp.nii'] ' -s ' 'freesurfer' ' -sd ' subdir ' -all'])
PAUSE POINT FreeSurfer’s fully automated segmentation and cortical extraction of a T1-weighted MRI can take 10 hours or more.
7) Import the extracted cortical surfaces into the MATLAB workspace and examine their quality. Repeat the following code using rh.pial to visualize the pial surface of the right hemisphere.
pial_lh = ft_read_headshape('freesurfer/surf/lh.pial');
pial_lh.coordsys = 'acpc';
ft_plot_mesh(pial_lh);
lighting gouraud;
camlight;
?TROUBLESHOOTING See the troubleshooting table in the original paper
8) Import the FreeSurfer-processed MRI into the MATLAB workspace for the purpose of fusing with the CT scan at a later step, and specify the coordinate system to which it was aligned in Step 4.
fsmri_acpc = ft_read_mri('freesurfer/mri/T1.mgz'); % on Windows, use 'SubjectUCI29_MR_acpc.nii'
fsmri_acpc.coordsys = 'acpc';
Preprocessing of the anatomical CT
9) Import the anatomical CT into the MATLAB workspace. Similar to the MRI, the CT scan comes in the format of a single file with an .img or .nii extension, or a folder containing a series of files with a .dcm or .ima extension (Supplementary File 2 may aid in the search and visualization of a DICOM series). For tutorial purposes, we read in the already preprocessed CT:
ct = ft_read_mri([subjID '_CT_acpc_f.nii']); % we used the dcm series
10) In case this cannot be done on the basis of knowledge of the laterality of electrode implantation, determine the native orientation of the anatomical CT’s left- right axis using ft_determine_coordsys, similarly to how it was done with the anatomical MRI in Step 3 (Box 3 and Supplementary Video 1).
CRITICAL STEP To correctly fuse the MRI and CT scans at a later step, accuracy in demarcating the right and left preauricular landmark in the following step is important for avoiding an otherwise hard to detect flip of the scan’s left and right orientation.
11) Align the anatomical CT to the CTF head surface coordinate system by specifying the nasion (at the root of the nose), left and right preauricular points (just in front of the ear canals), and an interhemispheric location along the midline at the top of the brain (Supplementary Video 3). The CT scan is initially aligned to the CTF head surface coordinate system given that the ACPC coordinate system used for the MRI relies on neuroanatomical landmarks that are not visible in the CT.
cfg = [];
cfg.method = 'interactive';
cfg.coordsys = 'ctf';
ct_ctf = ft_volumerealign(cfg, ct);
12) Automatically convert the CT’s coordinate system into an approximation of the ACPC coordinate system, the same system the anatomical MRI was aligned to. The call to ft_hastoolbox ensures that the required SPM toolbox is on the path.
ft_hastoolbox('spm12', 1);
ct_acpc = ft_convert_coordsys(ct_ctf, 'acpc');
Fusion of the CT with the MRI
13) Fuse the CT with the MRI, a necessary step to link the electrode locations in the anatomical CT to their corresponding locations in the anatomical MRI. Given that both scans are from the same subject and their common denominator is the skull, a rigid body transformation suffices for their alignment under normal circumstances (the default technique when using the SPM-method in FieldTrip).
cfg = [];
cfg.method = 'spm';
cfg.spmversion = 'spm12';
cfg.coordsys = 'acpc';
cfg.viewresult = 'yes';
ct_acpc_f = ft_volumerealign(cfg, ct_acpc, fsmri_acpc);
14) Carefully examine the interactive figure that is produced after the coregistration is completed, showing the MRI and fused CT superimposed. A successful fusion will show tight interlocking of CT-positive skull (in blue) and MRI-positive brain and skin tissue (in red).
CRITICAL STEP Accuracy of the fusion operation is important for correctly placing the electrodes in anatomical context in a following step.
?TROUBLESHOOTING
15) Write the MRI-fused anatomical CT out to file.
cfg = [];
cfg.filename = [subjID '_CT_acpc_f'];
cfg.filetype = 'nifti';
cfg.parameter = 'anatomy';
ft_volumewrite(cfg, ct_acpc_f);
Electrode placement
16) Import the header information from the recording file, if possible. By giving the electrode labels originating from the header as input to ft_electrodeplacement in the next step, the labels will appear as a to-do list during the interactive electrode placement activity. A second benefit is that the electrode locations can be directly assigned to labels collected from the recording file, obviating the need to sort and rename electrodes to match the electrophysiological data.
Raw recording files are not shared, in order to protect the subject’s identity. For tutorial purposes, load the header, which is normally obtained using ft_read_header, and continue with step 17:
load([subjID '_hdr.mat']);
hdr = ft_read_header(<path to recording file>);
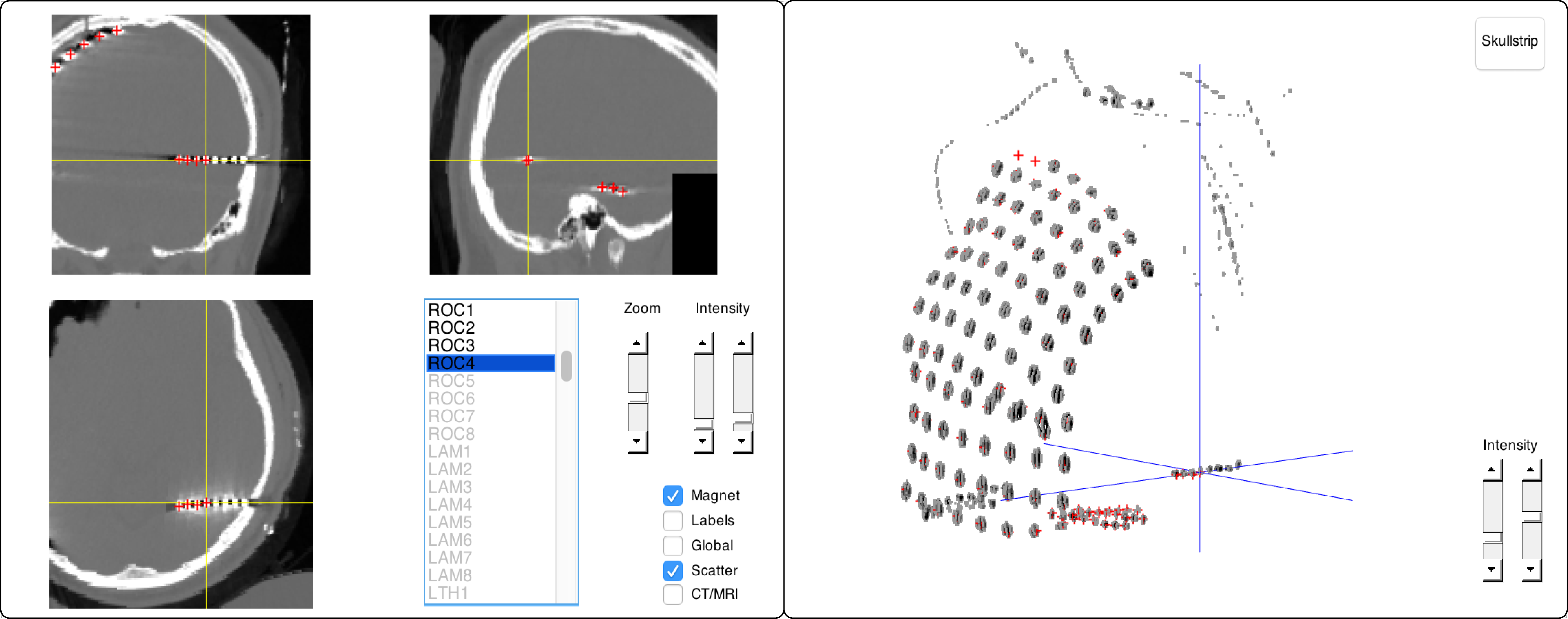
17) Localize the electrodes in the post-implant CT with ft_electrodeplacement, shown in the figure below. Clicking an electrode label in the list will directly assign that label to the current crosshair location (Supplementary Video 4). Several in-app features facilitate efficient yet precise navigation of the anatomical image, such as a zoom mode, a magnet option that transports the crosshair to the nearest weighted maximum with subvoxel accuracy (or minimum in case of a post-implant MRI), and an interactive three-dimensional scatter figure that is linked to the two-dimensional volume representations.
Furthermore, passing on the pre-implant MRI, fsmri_acpc, to ft_electrodeplacement allows toggling between CT and MRI views for the identification of specific electrodes based on their anatomical location. Generally, electrode #1 is the electrode farthest away from the craniotomy or burr hole in case of depths and single-row strips.
The present subject had 6 bilateral depths targeting left and right amygdalae (LAM/RAM) and the heads/anterior and tails/posterior of both hippocampi (LHH/RHH & LTH/RTH). Furthermore, one depth targeted right occipital cortex (ROC). Careful notes taken during surgery and recording are critical for determining the numbering of grid and multi-row strip electrodes. For an example, see this schematic drawing of the left parietal and temporal grids (LPG/LTG).
cfg = [];
cfg.channel = hdr.label;
elec_acpc_f = ft_electrodeplacement(cfg, ct_acpc_f, fsmri_acpc);
18) Examine whether the variables in resulting electrode structure elec_acpc_f match the recording parameters, e.g., the number of channels stored in the label field. The electrode and channel positions are stored in the elecpos and chanpos fields, respectively. The elecpos field contains the original electrode positions. With exception of possible brain shift compensation, this field is not adjusted.
The channel positions in the chanpos field are initially identical to the electrode positions but may be updated to accommodate offline adjustments in channel combinations, i.e. during re-montaging. For bipolar iEEG data, the best considered channel position is in between the two corresponding electrode positions. The chanpos field is used for overlaying the neural data on (sub-)cortical models during data visualization. The tra field is a matrix with the weight of each electrode into each channel, which at this stage merely is an identity matrix reflecting one-to-one mappings between electrodes and channels.
elec_acpc_f =
unit: 'mm'
coordsys: 'acpc'
label: {152x1 cell}
elecpos: [152x3 double]
chanpos: [152x3 double]
tra: [152x152 double]
cfg: [1x1 struct]
19) Visualize the MRI along with the electrodes and their labels and examine whether they show expected behavior.
figure
ft_plot_ortho(fsmri_acpc.anatomy, 'transform', fsmri_acpc.transform, 'style', 'intersect');
ft_plot_sens(elec_acpc_f, 'label', 'on', 'fontcolor', 'w', 'style', 'm.');
?TROUBLESHOOTING
20) Save the resulting electrode information to file.
save([subjID '_elec_acpc_f.mat'], 'elec_acpc_f');
Brain shift compensation (optional for cortical grids and strips)
21) In case of “brain shift”, i.e. the inward displacement of brain tissue and electrodes due to pressure changes related to the craniotomy, realignment of electrode grids to the pre-implant cortical surface may be necessary. To prevent inwardly displaced electrodes from being incorrectly placed in nearby cortical sulci during back-projection, create a smooth hull around the cortical mesh generated by FreeSurfer. This hull tracks the (exposed) outer surface on which the cortical grid rested.
The computation of the cortical hull takes some time. For tutorial purposes, load the hull from file and continue with step 23:
load([subjID '_hull_lh.mat']);
hull_lh = mesh;
cfg = [];
cfg.method = 'cortexhull';
cfg.headshape = 'freesurfer/surf/lh.pial';
cfg.fshome = <path to freesurfer home directory>; % for instance, '/Applications/freesurfer'
hull_lh = ft_prepare_mesh(cfg);
22) Save the hull to file.
save([subjID '_hull_lh.mat'], hull_lh);
23) Project the electrode grids to the surface hull of the implanted hemisphere. Given that different grids can move independently from one another and that the projection algorithm specified in cfg.warp considers the global electrode configuration of a grid, it is recommended to realign electrode grids individually by running separate realignment procedures for each grid. Here, we realign the electrodes of the left parietal grid followed by the electrodes of the left temporal grid (LPG and LTG respectively) and store the updated grid electrode information in a new variable together with the unaltered coordinates of the depth electrodes.
grids = {'LPG*', 'LTG*'};
elec_acpc_fr = elec_acpc_f; % start with a copy
for g = 1:numel(grids)
cfg = [];
cfg.channel = grids{g}; % first project LPGxx, then project LTGxx
cfg.keepchannel = 'yes';
cfg.elec = elec_acpc_fr;
cfg.method = 'headshape';
cfg.headshape = hull_lh;
cfg.warp = 'dykstra2012';
cfg.feedback = 'yes';
elec_acpc_fr = ft_electroderealign(cfg);
end
24) Visualize the cortex and electrodes together and examine whether they show expected behavior (figure below).
CRITICAL STEP Accuracy of the realignment operation is important for correctly placing the electrodes in anatomical context in a following step.
figure
ft_plot_mesh(pial_lh);
% ft_plot_mesh(hull_lh); % plot the hull
ft_plot_sens(elec_acpc_fr, 'style', 'k.');
% ft_plot_sens(elec_acpc_f, 'style', 'r.'); % plot the electrodes prior to projection
view([-55 10])
material dull
% alpha 0.5 % make the surface mesh transparent
lighting gouraud
camlight
?TROUBLESHOOTING
25) Save the updated electrode information to file.
save([subjID '_elec_acpc_fr.mat'], 'elec_acpc_fr');
Volume-based registration (optional)
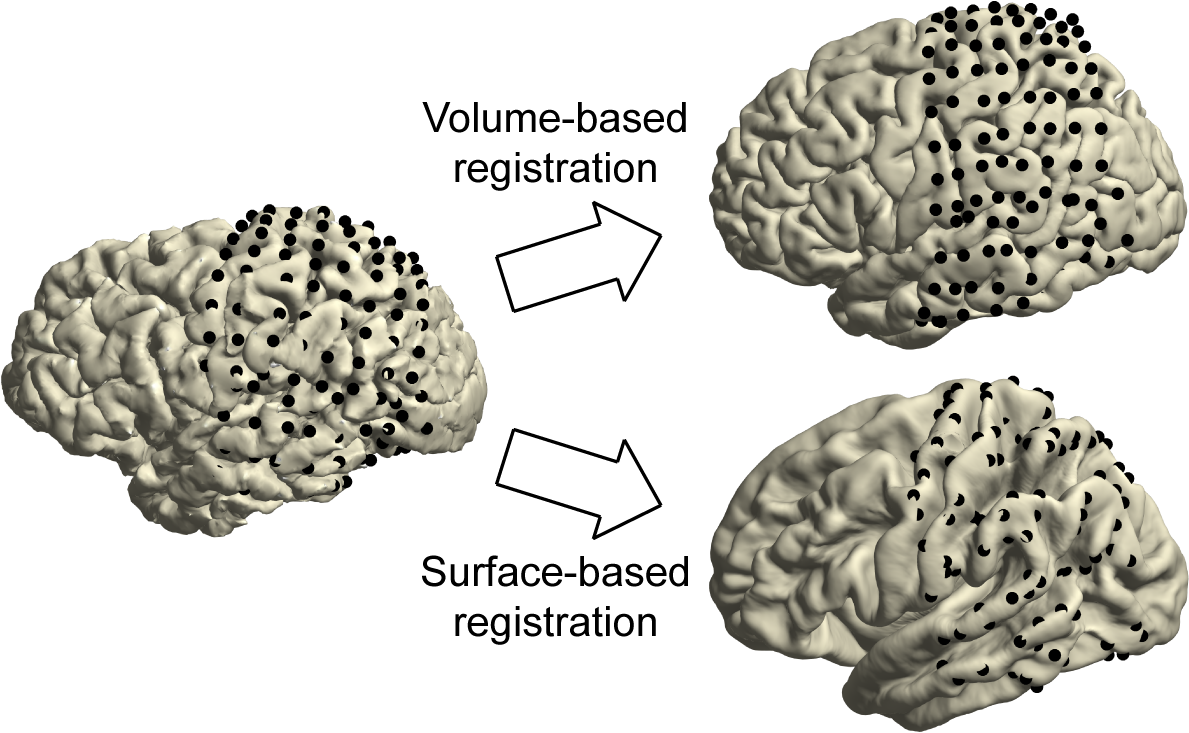
26 & 27) To generalize the electrode coordinates to other brains or MNI-based neuroanatomical atlases in a later step, register the subject’s brain to the standard MNI brain and use the resulting deformation parameters to obtain the electrodes in standard MNI space. The volume-based registration technique considers the overall geometry of the brain and can be used for the spatial normalization of all types of electrodes, whether depth or on the surface.
cfg = [];
cfg.elec = elec_acpc_fr;
cfg.method = 'mni';
cfg.mri = fsmri_acpc;
cfg.spmversion = 'spm12';
cfg.spmmethod = 'new';
cfg.nonlinear = 'yes';
elec_mni_frv = ft_electroderealign(cfg);
28) Visualize the cortical mesh extracted from the standard MNI brain along with the spatially normalized electrodes and examine whether they show expected behavior given the electrodes’ original anatomical locations (top right in the figure below).
CRITICAL STEP Accuracy of the spatial normalization step is important for correctly overlaying the electrode positions with a brain atlas in a following step.
[ftver, ftpath] = ft_version;
load([ftpath filesep 'template/anatomy/surface_pial_left.mat']);
% rename the variable that we read from the file, as not to confuse it with the MATLAB mesh plotting function
template_lh = mesh; clear mesh;
ft_plot_mesh(template_lh);
ft_plot_sens(elec_mni_frv);
view([-90 20])
material dull
lighting gouraud
camlight
?TROUBLESHOOTING
29) Save the normalized electrode information to file.
save([subjID '_elec_mni_frv.mat'], 'elec_mni_frv');
Surface-based registration (optional for surface electrodes)
30) To generalize the electrode coordinates to other brains in a later step, map the electrodes onto FreeSurfer’s fsaverage brain. The surface-based registration technique solely considers the curvature patterns of the cortex and thus can be used for the spatial normalization of electrodes located on or near the cortical surface. In the example case, this pertains to all electrodes of the left parietal and temporal grids.
cfg = [];
cfg.elec = elec_acpc_fr;
cfg.channel = {'LPG*', 'LTG*'};
cfg.method = 'headshape';
cfg.headshape = 'freesurfer/surf/lh.pial';
cfg.warp = 'fsaverage';
cfg.fshome = <path to freesurfer home directory>; % for instance, '/Applications/freesurfer'
elec_fsavg_frs = ft_electroderealign(cfg);
31) Visualize FreeSurfer’s fsaverage brain along with the spatially normalized electrodes and examine whether they show expected behavior (bottom right in the figure below).
CRITICAL STEP Accuracy of the spatial normalization step is important for correctly overlaying the electrode positions with a brain atlas in a following step.
fspial_lh = ft_read_headshape([cfg.fshome '/subjects/fsaverage/surf/lh.pial']);
fspial_lh.coordsys = 'fsaverage';
ft_plot_mesh(fspial_lh);
ft_plot_sens(elec_fsavg_frs);
view([-90 20])
material dull
lighting gouraud
camlight
32) Save the normalized electrode information to file.
save([subjID '_elec_fsavg_frs.mat'], 'elec_fsavg_frs');
Anatomical labeling (optional)
33) FieldTrip supports looking up the anatomical or functional labels corresponding to the electrodes in a number of atlases, including the AFNI Talairach Tournoux atlas, the AAL atlas, the BrainWeb data set, the JuBrain cytoarchitectonic atlas, the VTPM atlas, and the Brainnetome atlas, in addition to the subject-tailored Desikan-Killiany and Destrieux atlases produced by FreeSurfer. Given that no two electrodes end up in the exact same location across subjects due to inter-individual variability in electrode coverage and brain anatomy, atlases are particularly useful for the systematic combination of neural activity from different subjects in a so-called region of interest (ROI) analysis. With exception of the above FreeSurfer-based atlases, the atlases are in MNI coordinate space and require the electrodes to be spatially normalized (Steps 26 through 27). First, import an atlas of interest, e.g., the AAL atlas, into the MATLAB workspace.
[ftver, ftpath] = ft_version;
atlas = ft_read_atlas([ftpath filesep 'template/atlas/aal/ROI_MNI_V4.nii']);
34) Look up the corresponding anatomical label of an electrode of interest, e.g., electrode LHH1, targeting the left hemisphere’s hippocampus. Supplementary File 3 represents a tool that automatically overlays all channels in an electrode structure with all of the above atlases and stores the resulting anatomical labels in an excel table (e.g., SubjectUCI29_electable.xlsx in the zip file). A more recent version of this tool can be found here.
cfg = [];
cfg.roi = elec_mni_frv.chanpos(match_str(elec_mni_frv.label,'LHH1'),:);
cfg.atlas = atlas;
cfg.output = 'single';
labels = ft_volumelookup(cfg, atlas);
[~, indx] = max(labels.count);
labels.name(indx)
ans =
'ParaHippocampal_L'
?TROUBLESHOOTING
Functional workflow
Preprocessing of the neural recordings
35) Define the trials, that is, the segments of data that will be used for further processing and analysis. This step produces a matrix cfg.trl containing for each segment the begin and end sample in the recording file. In the case of the example provided in the shared data, the segments of interest begin 400 ms before tone onset, are marked with a ‘4’ in the trigger channel, and end 900 ms thereafter.
Raw recording files are not shared, in order to protect the subject’s identity. For tutorial purposes, load the preprocessed data, which is the product of steps 35 & 36, and continue with step 37:
load([subjID '_data.mat'], 'data');
cfg = [];
cfg.dataset = <path to recording file>;
cfg.trialdef.eventtype = 'TRIGGER';
cfg.trialdef.eventvalue = 4;
cfg.trialdef.prestim = 0.4;
cfg.trialdef.poststim = 0.9;
cfg = ft_definetrial(cfg);
36) Import the data segments of interest into the MATLAB workspace and filter the data for high-frequency and power line noise (see the documentation of ft_preprocessing for filtering options).
cfg.demean = 'yes';
cfg.baselinewindow = 'all';
cfg.lpfilter = 'yes';
cfg.lpfreq = 200;
cfg.padding = 2;
cfg.padtype = 'data';
cfg.bsfilter = 'yes';
cfg.bsfiltord = 3;
cfg.bsfreq = [59 61; 119 121; 179 181]; % line noise at 60 Hz in USA
data = ft_preprocessing(cfg);
37) Examine whether the variables in the output data structure match the recording and preprocessing parameters, i.e. the sampling rate (fsample), number of recording channels (label), and segmentation into the experiment’s twenty-six trials (trial, and their respective time axes in time).
data =
time: {1x26 cell}
label: {152x1 cell}
trial: {1x26 cell}
fsample: 5000
sampleinfo: [26x2 double]
trialinfo: [26x1 double]
38) Add the elec structure originating from the anatomical workflow and save the preprocessed electrophysiological data to file. The advantage of adding the electrode information at this stage is that it will be kept consistent with the neural data going forward, as when applying the same montage used for the neural recordings to the channel positions.
data.elec = elec_acpc_fr;
save([subjID '_data.mat'], 'data');
39) Inspect the neural recordings using ft_databrowser and identify channels or segments of non-interest, for instance segments containing signal artifacts or (in this case) epileptiform activity. Mark the bad segments by drawing a box around the corrupted signal. Write down the labels of bad channels.
CRITICAL STEP Identifying bad channels is important for avoiding the contamination of other channels during re-montaging in Step 41.
cfg = [];
cfg.viewmode = 'vertical';
cfg = ft_databrowser(cfg, data);
40) Remove any bad segments marked in the above step.
cfg.artfctdef.reject = 'complete';
data = ft_rejectartifact(cfg, data);
41) Re-montage the cortical grids to a common average reference in order to remove noise that is shared across all channels. Box 4 provides a background on re-montaging. Bad channels identified in Step 39 can be excluded from this step by adding those channels to cfg.channel with a minus prefix. That is, cfg.channel = {‘LPG’, ‘LTG’, ‘-LPG1’} if one were to exclude the LPG1 channel from the list of LPG and LTG channels.
cfg = [];
cfg.channel = {'LPG*', 'LTG*'};
cfg.reref = 'yes';
cfg.refchannel = 'all';
cfg.refmethod = 'avg';
reref_grids = ft_preprocessing(cfg, data);
42) Apply a bipolar montage to the depth electrodes. This can be done in a similar manner as in Step 41, but by selecting single channel labels for cfg.channel and cfg.refchannel. As shown below, it can also be done using cfg.refmethod = ‘bipolar’, which automatically takes bipolar derivations of consecutive channels. New channel labels in the output indicate the bipolar origin of the data, e.g., “RAM1-RAM2”, “RAM2-RAM3”, and so on. By specifying cfg.updatesens = ‘yes’, the same bipolar montage is automatically applied to the electrode positions, with the resulting chanpos field containing the mean locations of the electrode pair that comprises a bipolar channel. More elaborate schemes can be created by the specification of weight matrices using cfg.montage that define how existing channels should be combined into new channels (see the documentation of ft_apply_montage).
depths = {'RAM*', 'RHH*', 'RTH*', 'ROC*', 'LAM*', 'LHH*', 'LTH*'};
for d = 1:numel(depths)
cfg = [];
cfg.channel = ft_channelselection(depths{d}, data.label);
cfg.reref = 'yes';
cfg.refchannel = 'all';
cfg.refmethod = 'bipolar';
cfg.updatesens = 'yes';
reref_depths{d} = ft_preprocessing(cfg, data);
end
43) Combine the data from both electrode types into one data structure for the ease of further processing.
cfg = [];
cfg.appendsens = 'yes';
reref = ft_appenddata(cfg, reref_grids, reref_depths{:});
44) Save the re-referenced data to file.
save([subjID '_reref.mat'], reref);
Time-frequency analysis (optional)
45) Decompose the signal in time and frequency bins using time-resolved Fourier-based spectral decomposition. The settings for spectral decomposition depend on the clinical or research question at hand and contingencies in the experimental paradigm. Given that we plan to look at task-related changes in high-frequency-band activity (70 to 150 Hz) in a following step, we sample the neural activity using cfg.toi throughout the 300 ms baseline period before tone onset at 0 ms and until after the approximate button press time at 700 ms. At each time point, we estimate spectral content using 200 ms windows (cfg.t_ftimwin). This time window captures 20 cycles of a 100 Hz rhythm, which is well over the 3 cycles minimally required to unambiguously recover the spectral information. For a broader view of powerspectral modulations, we extend the frequency range using cfg.foi from 5 to 200 Hz. Finally, note that cfg.keeptrials is set to ‘no’ here (as it is by default), meaning that the average spectral content across all trials is returned (see the documentation of ft_freqanalysis).
cfg = [];
cfg.method = 'mtmconvol';
cfg.toi = -.3:0.01:.8;
cfg.foi = 5:5:200;
cfg.t_ftimwin = ones(length(cfg.foi),1).*0.2;
cfg.taper = 'hanning';
cfg.output = 'pow';
cfg.keeptrials = 'no';
freq = ft_freqanalysis(cfg, reref);
46) Save the time-frequency data to file.
save([subjID '_freq.mat'], 'freq');
Interactive plotting
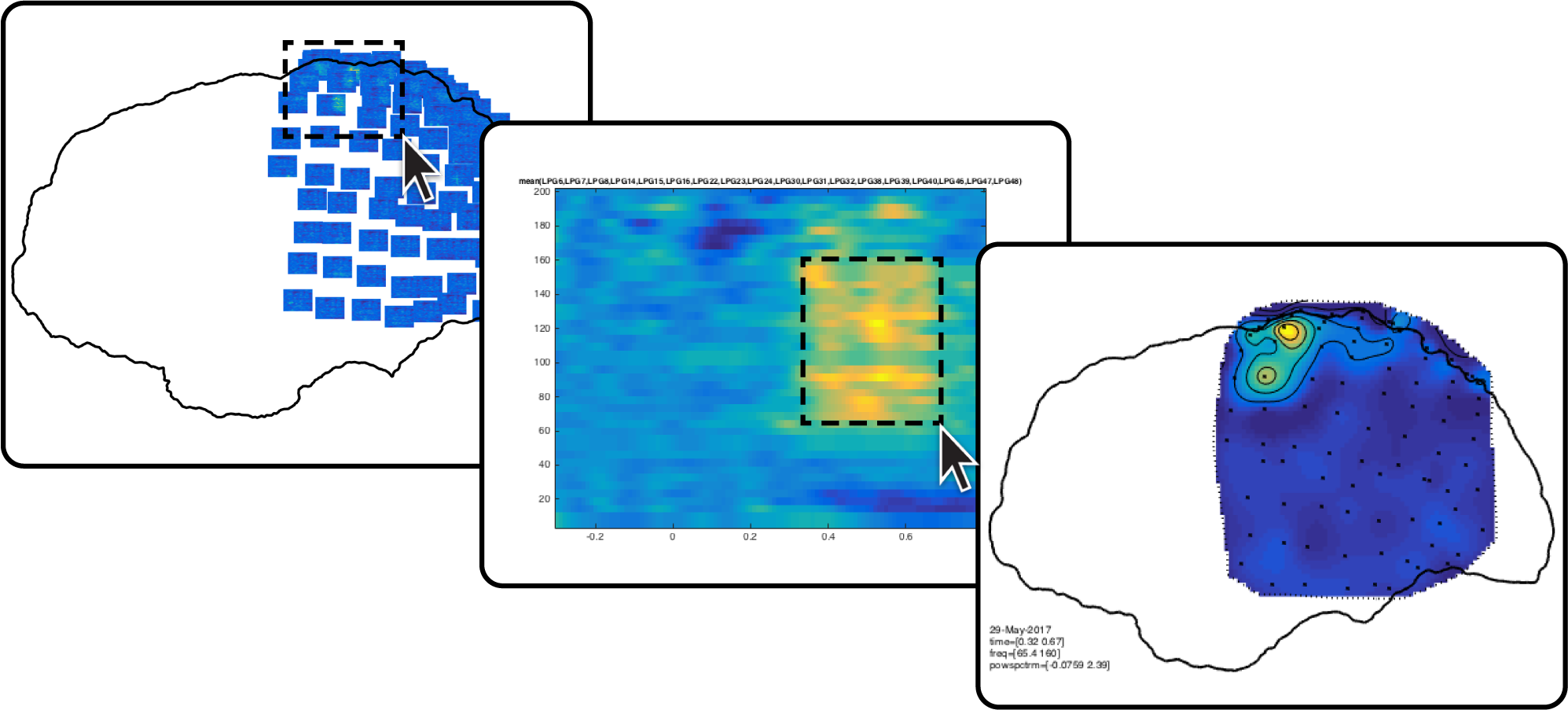
47) For an anatomically informed exploration of the multidimensional outcome of an analysis, create a layout based on the three-dimensional electrode locations. This layout is a symbolic representation in which the channels are projected on the two-dimensional medium offered by paper or a computer screen, see also the layout tutorial.
The layout is complemented by an automatic outline of the cortical sheet that is specified in cfg.headshape. The cfg.boxchannel option allows selecting channels whose two-dimensional distances are used to determine the plotting box sizes in the following step.
cfg = [];
cfg.headshape = pial_lh;
cfg.projection = 'orthographic';
cfg.channel = {'LPG*', 'LTG*'};
cfg.viewpoint = 'left';
cfg.mask = 'convex';
cfg.boxchannel = {'LTG30', 'LTG31'};
cfg.feedback = 'yes';
lay = ft_prepare_layout(cfg, freq);
48) Express the time-frequency representation of neural activity at each channel in terms of the relative change in activity from the baseline interval. We exclude the baseline interval from -100 to 0 ms given that powerspectral data points in this interval were estimated using 200 ms time windows that extend into the task period.
cfg = [];
cfg.baseline = [-.3 -.1];
cfg.baselinetype = 'relchange';
freq_blc = ft_freqbaseline(cfg, freq);
49) Visualize the time-frequency representations overlaid on the two-dimensional layout. The generated figure is interactive, so that selecting a group of channels will launch another figure representing the average time-frequency representation over those channels (figure below). Selecting a certain frequency and time range in that time-frequency representation will launch yet another figure showing the topographical distribution of activity in the selected interval, and so on (Supplementary Video 5).
cfg = [];
cfg.layout = lay;
cfg.showoutline = 'yes';
ft_multiplotTFR(cfg, freq_blc);
ECoG data representation
50) For an anatomically realistic representation of cortical activity, overlay a surface model of the neocortex with the spatial distribution of the high frequency-band activity. First, extract mean high-frequency-band activity during a time interval of interest.
cfg = [];
cfg.frequency = [70 150];
cfg.avgoverfreq = 'yes';
cfg.latency = [0 0.8];
cfg.avgovertime = 'yes';
freq_sel = ft_selectdata(cfg, freq_blc);
51) Visualize the spatial distribution of high-frequency-band activity on a cortical mesh of the subject’s brain.
cfg = [];
cfg.funparameter = 'powspctrm';
cfg.funcolorlim = [-.5 .5];
cfg.method = 'surface';
cfg.interpmethod = 'sphere_weighteddistance';
cfg.sphereradius = 8;
cfg.camlight = 'no';
ft_sourceplot(cfg, freq_sel, pial_lh);
view([-90 20])
material dull
lighting gouraud
camlight
52) Add the electrodes to the figure (shown below). By looping around Steps 50 and 51 while breaking down the time interval of interest specified with cfg.latency in consecutive steps, it becomes feasible to observe the spatiotemporal dynamics of neural activity occurring in relation to known experimental structure and behavior (Supplementary Video 6). See help getframe for capturing and assembling time-lapse movies.
ft_plot_sens(elec_acpc_fr);
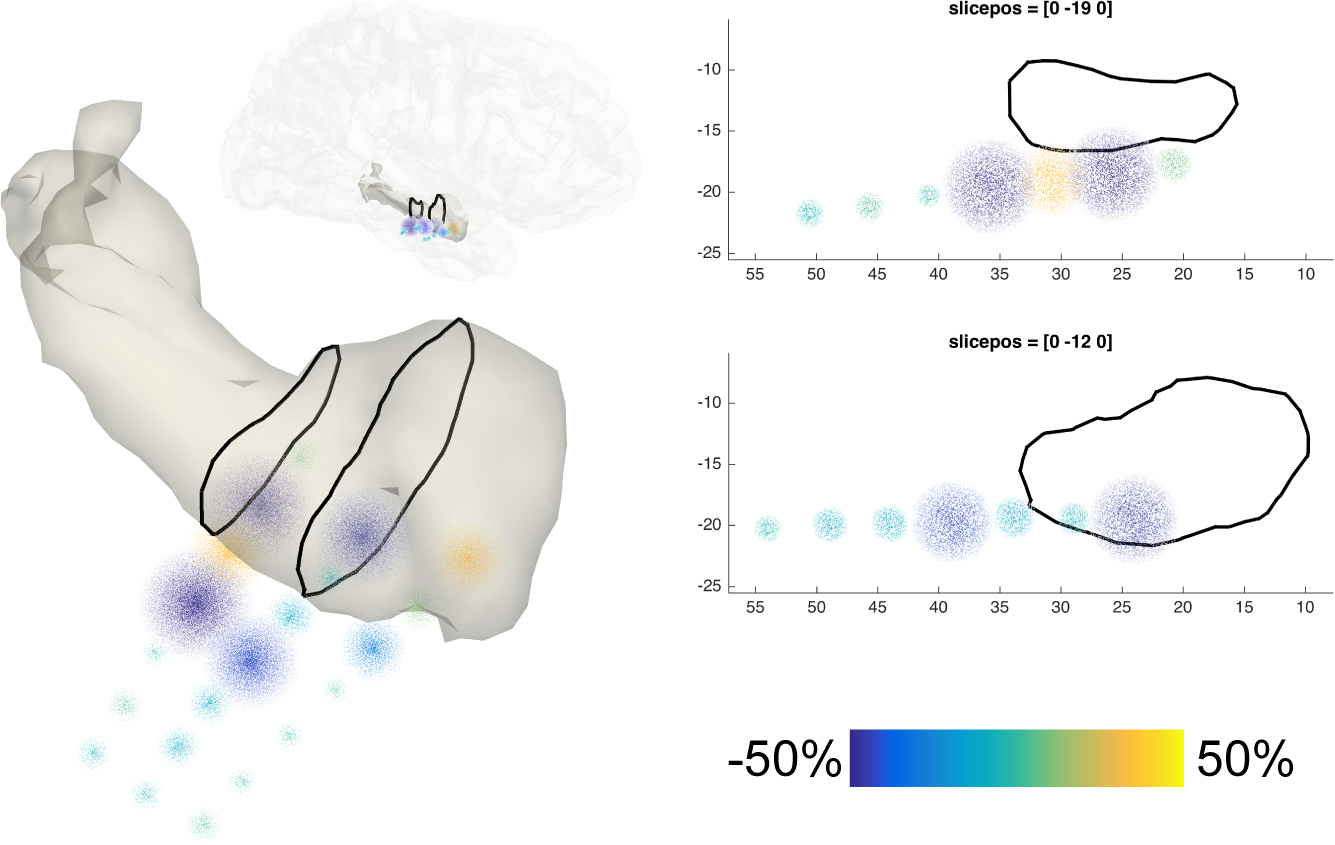
SEEG data representation
53) For depth recordings, create an integrated representation of neural activity and anatomy by interpolating neural data from each bipolar channel in a spherical cloud, which can then be overlaid on a surface mesh of any deep brain structure. First, create a volumetric mask of the regions of interest. Here, we generate a mask for the right hippocampus and amygdala from the cortical parcellation and subcortical segmentation produced by FreeSurfer.
atlas = ft_read_atlas('freesurfer/mri/aparc+aseg.mgz');
atlas.coordsys = 'acpc';
cfg = [];
cfg.atlas = atlas;
cfg.roi = {'Right-Hippocampus', 'Right-Amygdala'};
mask_rha = ft_volumelookup(cfg, atlas);
54) Create a triangulated and smoothed surface mesh on the basis of the volumetric masks.
seg = keepfields(atlas, {'dim', 'unit','coordsys','transform'});
seg.brain = mask_rha;
cfg = [];
cfg.method = 'iso2mesh';
cfg.radbound = 2;
cfg.maxsurf = 0;
cfg.tissue = 'brain';
cfg.numvertices = 1000;
cfg.smooth = 3;
cfg.spmversion = 'spm12';
mesh_rha = ft_prepare_mesh(cfg, seg);
55) Identify the subcortical electrodes of interest.
cfg = [];
cfg.channel = {'RAM*', 'RTH*', 'RHH*'};
freq_sel2 = ft_selectdata(cfg, freq_sel);
56) Interpolate the high-frequency-band activity in the bipolar channels on a spherical cloud around the channel positions, while overlaying the neural activity with the above mesh (Supplementary File 4 highlights the currently available cloud types for plotting). By repeating the current step for neural data corresponding to consecutive time intervals, similarly to the process outlined in Step 52, it becomes feasible to create time-lapse movies of the spatiotemporal dynamics of deep-brain activity (Supplementary Video 7 shows the spatiotemporal evolution of epileptiform activity in a separate subject).
cfg = [];
cfg.method = 'cloud';
cfg.funparameter = 'powspctrm';
cfg.funcolorlim = [-.5 .5];
cfg.slice = '3d';
cfg.nslices = 2;
cfg.facealpha = .25;
ft_sourceplot(cfg, freq_sel2, mesh_rha);
view([120 40])
lighting gouraud
camlight
57) To create a more definitive image of the neural activity at particular positions, generate two-dimensional slices through the three-dimensional representations. This combination provides the most complete and integrated representation of neural and anatomical data (figure below).
cfg.slice = '2d';
ft_sourceplot(cfg, freq_sel2, mesh_rha);
Summary and conclusion
Upon completion of this tutorial, one should obtain an integrated representation of neural and anatomical data. The exact results depend ultimately on the clinical or research question at hand, contingencies in the experimental paradigm, and decisions made during the execution of the protocol.
This tutorial demonstrated the analysis of spatiotemporal neural dynamics occurring in relation to known experimental structure and relatively simple behavior, namely the pressing of a button with the right hand when hearing a target tone. However, with small adaptations of the protocol it is feasible to track the spatiotemporal evolution of epileptiform activity with high precision (e.g., Supplementary Video 7), or to perform group-level investigations of fine-grained emotion- or language-related neural dynamics in human hippocampus. A precise fusion of the anatomical images with the electrophysiological data is key to reproducible analyses and findings. Hence, it is important to examine the outcome of any critical step, as we have done above.
We recommend that the user construct a single script for a single subject by copying and pasting code from this protocol into the MATLAB editor (e.g., Supplementary File 1), and evaluating segments of that script in the MATLAB command window. Once the script produces satisfactory results, it can be converted into a batch analysis by breaking it into separate components. By looping around the separate components for all subjects, the entire analysis pipeline for all subjects in a study can easily be executed and intermediate results can be saved and evaluated.
Suggested further reading
You can read more about the challenges and concepts of preprocessing intracranial EEG data in the Intracranial EEG for Cognitive Neuroscience handbook. You can read more about other types of intracranial recordings such as spike train recordings and spikes and local field potentials in the following documentation.
- Analysis of monkey ECoG recordings
- Channel and source analysis of mouse EEG
- Preprocessing and analysis of spike-train data
- Preprocessing and analysis of spike and local field potential data
Here is also a list of related documentation:
FAQs on data formats
- How can I append the files of two separate recordings?
- What dataformats are supported?
- How are the various MATLAB data structures defined?
- How can I convert an anatomical MRI from DICOM into CTF format?
- How can I convert one dataformat into an other?
- How can I extend the reading functions with a new dataformat?
- How can I import my own data format?
- How can I merge two datasets that were acquired simultaneously with different amplifiers?
- I have problems reading in neuroscan .cnt files. How can I fix this?
- Reading is slow, can I write my raw data to a more efficient file format?
FAQs on frequency analysis
- Does it make sense to subtract the ERP prior to time frequency analysis, to distinguish evoked from induced power?
- How can I do time-frequency analysis on continuous data?
- How can I test whether a behavioral measure is phasic?
- In what way can frequency domain data be represented in FieldTrip?
- How does MTMCONVOL work?
- What are the differences between the old and the new implementation of 'mtmconvol' in ft_freqanalyis?
- What are the differences between the old and the new implementation of 'mtmftt' in ft_freqanalyis?
- What are the differences between the old and the new implementation of 'wavelet' (formerly 'wltconvol') in ft_freqanalyis?
- What convention is used to define absolute phase in 'mtmconvol', 'wavelet' and 'mtmfft'?
- What does "padding not sufficient for requested frequency resolution" mean?
- What kind of filters can I apply to my data?
- Why am I not getting integer frequencies?
- Why does my output.freq not match my cfg.foi when using 'mtmconvol' in ft_freqanalyis?
- Why does my output.freq not match my cfg.foi when using 'mtmfft' in ft_freqanalyis
- Why does my output.freq not match my cfg.foi when using 'wavelet' (formerly 'wltconvol') in ft_freqanalyis?
- Why does my TFR contain NaNs?
- Why does my TFR look strange (part I, demeaning)?
- Why does my TFR look strange (part II, detrending)?
- Why is the largest peak in the spectrum at the frequency which is 1/segment length?