workshop / meg-uk-2015 / fieldtrip-connectivity-demo /
FieldTrip connectivity demo
In this demonstration we will use the face recognition dataset.
Please use the general instructions to get started.
Part 1 - virtual channel connectivity
%% start with data that was preprocessed in FieldTrip
subj = 15;
prefix = sprintf('Sub%02d', subj);
load([prefix '_raw']); % this is called "data" rather than "raw"
% load the results from beamformer_part1
load vol
load sens
load mri_realigned
%% deal with maxfilter
% the data has been maxfiltered and subsequently contatenated
% this results in an ill-conditioned estimate of covariance or CSD
cfg = [];
cfg.method = 'pca';
cfg.updatesens = 'no';
cfg.channel = 'MEGMAG';
comp = ft_componentanalysis(cfg, data);
cfg = [];
cfg.updatesens = 'no';
cfg.component = comp.label(51:end);
data_fix = ft_rejectcomponent(cfg, comp);
%%
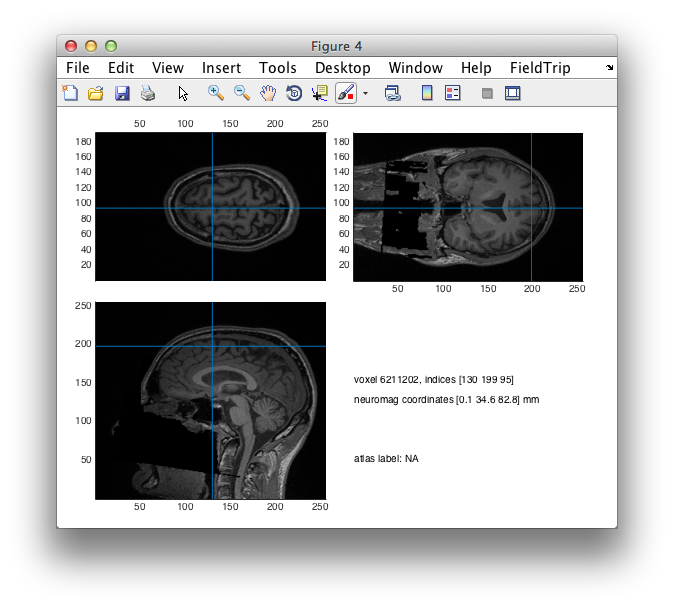
pos1 = [21 -64 30];
pos2 = [0 35 83];
cfg = [];
cfg.location = pos1;
figure; ft_sourceplot(cfg, mri_realigned);
cfg.location = pos2;
figure; ft_sourceplot(cfg, mri_realigned);
%%
% timelock2 was computed in https://www.fieldtriptoolbox.org/workshop/meg-uk-2015/fieldtrip-beamformer-demo#part_3_-_reconstruct_single-trial_cortical_responses
cfg = [];
cfg.headmodel = vol;
cfg.grad = sens;
cfg.senstype = 'meg';
cfg.method = 'lcmv';
cfg.lcmv.keepfilter = 'yes';
cfg.lcmv.projectmom = 'yes';
cfg.unit = 'mm';
cfg.sourcemodel.pos = pos1;
source1 = ft_sourceanalysis(cfg, timelock2);
cfg.sourcemodel.pos = pos2;
source2 = ft_sourceanalysis(cfg, timelock2);
%% construct single-trial virtual channel representation
virtualchannel_raw = [];
virtualchannel_raw.label = {'pos1'; 'pos2'};
virtualchannel_raw.trialinfo = data_fix.trialinfo;
for i=1:882
% note that this is the non-filtered raw data
virtualchannel_raw.time{i} = data_fix.time{i};
virtualchannel_raw.trial{i}(1,:) = source1.avg.filter{1} * data_fix.trial{i}(:,:);
virtualchannel_raw.trial{i}(2,:) = source2.avg.filter{1} * data_fix.trial{i}(:,:);
end
%%
cfg = [];
cfg.keeptrials = 'yes';
cfg.preproc.demean = 'yes';
cfg.preproc.baselinewindow = [-inf 0];
virtualchannel_avg = ft_timelockanalysis(cfg, virtualchannel_raw);
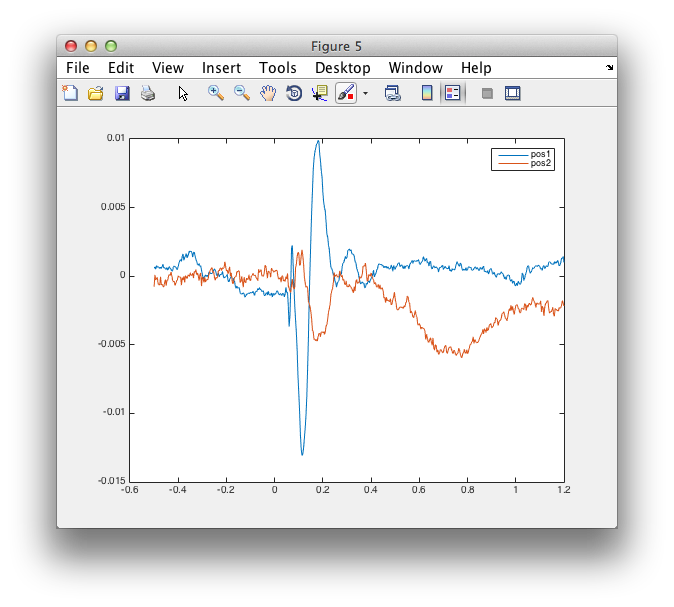
figure
plot(virtualchannel_avg.time, virtualchannel_avg.avg)
legend(virtualchannel_avg.label);
%%
cfg = [];
cfg.method = 'wavelet';
cfg.output = 'powandcsd';
cfg.foi = 4:2:70;
cfg.toi = -0.200:0.020:1.000;
virtualchannel_wavelet = ft_freqanalysis(cfg, virtualchannel_raw);
cfg = [];
cfg.baselinetype = 'relative';
cfg.baseline = [-inf 0];
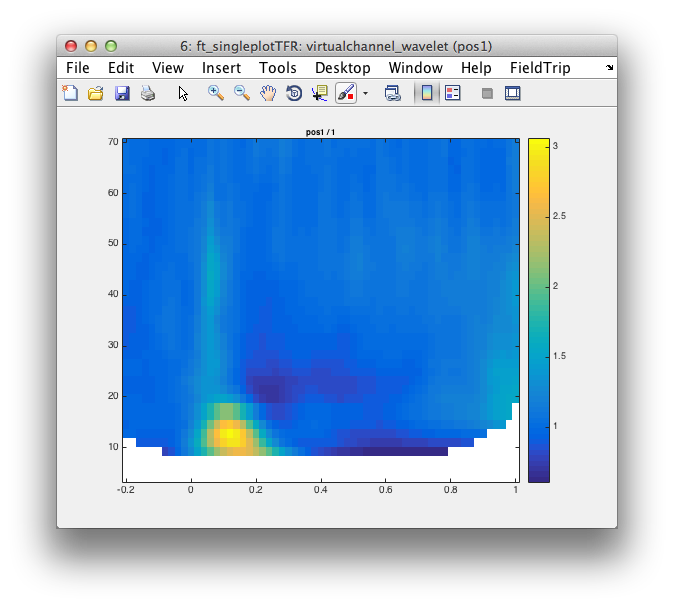
cfg.channel = {'pos1'};
figure; ft_singleplotTFR(cfg, virtualchannel_wavelet);
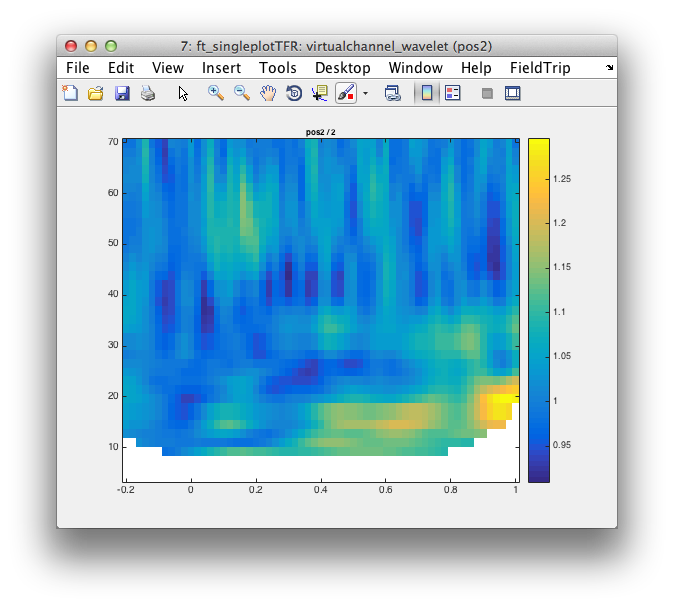
cfg.channel = {'pos2'};
figure; ft_singleplotTFR(cfg, virtualchannel_wavelet);
%%
cfg = [];
cfg.method = 'coh';
coherence = ft_connectivityanalysis(cfg, virtualchannel_wavelet);
figure
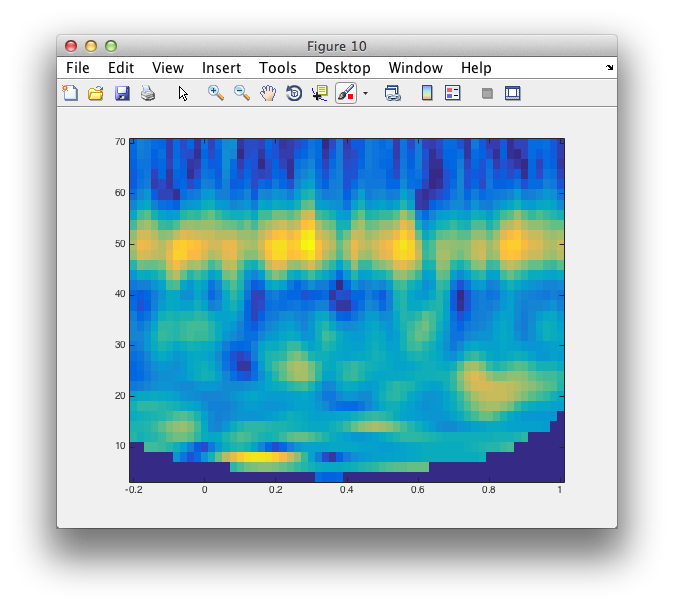
imagesc(coherence.time, coherence.freq, squeeze(coherence.cohspctrm(1,:,:)));
axis xy
Part 2 - whole brain connectivity
%% start from data that was processed by FieldTrip
subj = 15;
prefix = sprintf('Sub%02d', subj);
load([prefix '_raw']); % this is called "data" rather than "raw"
% load the results from beamformer_part1
load vol
load sens
load mri_realigned
%% deal with maxfilter
% the data has been maxfiltered and subsequently contatenated
% this results in an ill-conditioned estimate of covariance or CSD
cfg = [];
cfg.method = 'pca';
cfg.updatesens = 'no';
cfg.channel = 'MEGMAG';
comp = ft_componentanalysis(cfg, data);
cfg = [];
cfg.updatesens = 'no';
cfg.component = comp.label(51:end);
data_fix = ft_rejectcomponent(cfg, comp);
%%
cfg = [];
cfg.channel = 'MEGMAG';
cfg.method = 'wavelet';
cfg.output = 'fourier';
cfg.keeptrials = 'yes';
cfg.foi = 16;
cfg.toi = 0.150;
freq = ft_freqanalysis(cfg, data_fix);
%%
cfg = [];
cfg.resolution = 7;
% cfg.inwardshift = -7; % allow dipoles 10mm outside the brain, this improves interpolation at the edges
cfg.unit = 'mm';
cfg.headmodel = vol; % from FT
cfg.grad = sens; % from FT
cfg.senstype = 'meg';
cfg.normalize = 'yes';
grid = ft_prepare_leadfield(cfg, freq);
% save grid grid
%%
cfg = [];
cfg.headmodel = vol; % from FT
cfg.grad = sens; % from FT
cfg.senstype = 'meg';
cfg.grid = grid;
cfg.method = 'pcc';
cfg.pcc.fixedori = 'yes';
cfg.latency = [0.140 0.160];
cfg.frequency = [14 18];
source = ft_sourceanalysis(cfg, freq);
figure
plot(source.avg.mom{source.inside(1)}, '.')
xlabel('real');
ylabel('imag');
%%
pos = [21 -64 30];
% compute the nearest grid location
dif = grid.pos;
dif(:,1) = dif(:,1)-pos(1);
dif(:,2) = dif(:,2)-pos(2);
dif(:,3) = dif(:,3)-pos(3);
dif = sqrt(sum(dif.^2,2));
[distance, refindx] = min(dif);
cfg = [];
cfg.method = 'coh';
% cfg.complex = 'abs';
cfg.complex = 'absimag';
cfg.refindx = refindx;
conn = ft_connectivityanalysis(cfg, source);
% the output contains both the actual source position, as well as the position of the reference
% this is ugly and will probably change in future FieldTrip versions
orgpos = conn.pos(:,1:3);
refpos = conn.pos(:,4:6);
conn.pos = orgpos;
%% visualize the seed-based connectivity results
cfg = [];
cfg.funparameter = 'cohspctrm';
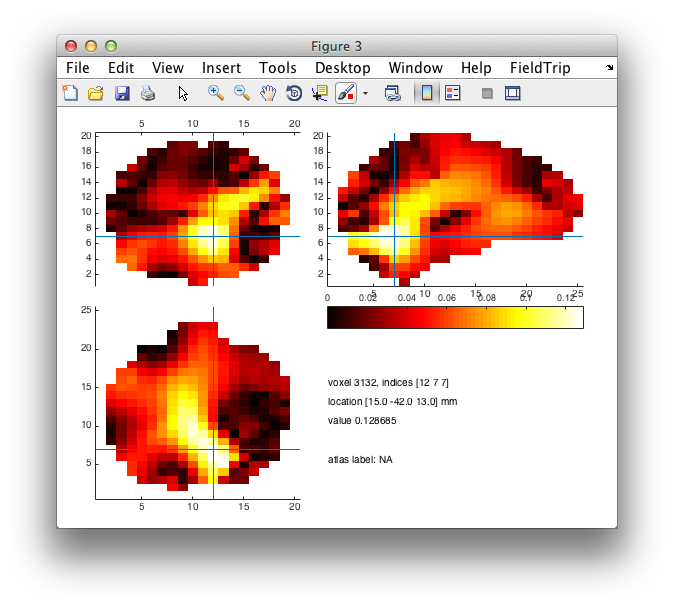
figure; ft_sourceplot(cfg, conn);
cfg = [];
cfg.parameter = 'cohspctrm';
sourceI = ft_sourceinterpolate(cfg, conn, mri_realigned);
cfg = [];
cfg.funparameter = 'cohspctrm';
figure; ft_sourceplot(cfg, sourceI);
%% look at connectivity difference
cfg = [];
cfg.trials = find(freq.trialinfo==1 | freq.trialinfo==2); % 1=Famous, 2=Unfamiliar
source1 = ft_selectdata(cfg, source);
cfg.trials = find(freq.trialinfo==3); % 3=Scrambled
source2 = ft_selectdata(cfg, source);
% consider using ft_stratify to equalise number of trials going in to source 1 and 2, as SNR differences between conditions affects connectivity measures between conditions
cfg = [];
cfg.method = 'coh';
cfg.complex = 'absimag';
cfg.refindx = refindx;
conn1 = ft_connectivityanalysis(cfg, source1);
conn2 = ft_connectivityanalysis(cfg, source2);
conn1.pos = conn1.pos(:,1:3);
conn2.pos = conn2.pos(:,1:3);
cfg = [];
cfg.parameter = 'cohspctrm';
cfg.operation = 'x1-x2'; % or 'subtract'
conn_dif = ft_math(cfg, conn1, conn2);
cfg = [];
cfg.parameter = 'cohspctrm';
source1int = ft_sourceinterpolate(cfg, conn1, mri_realigned);
source2int = ft_sourceinterpolate(cfg, conn2, mri_realigned);
source_difint = ft_sourceinterpolate(cfg, conn_dif, mri_realigned);
cfg = [];
cfg.funparameter = 'cohspctrm';
cfg.funcolorlim = [-0.1 0.1];
cfg.maskparameter = 'cohspctrm';
cfg.opacitylim = [-0.15 0.15];
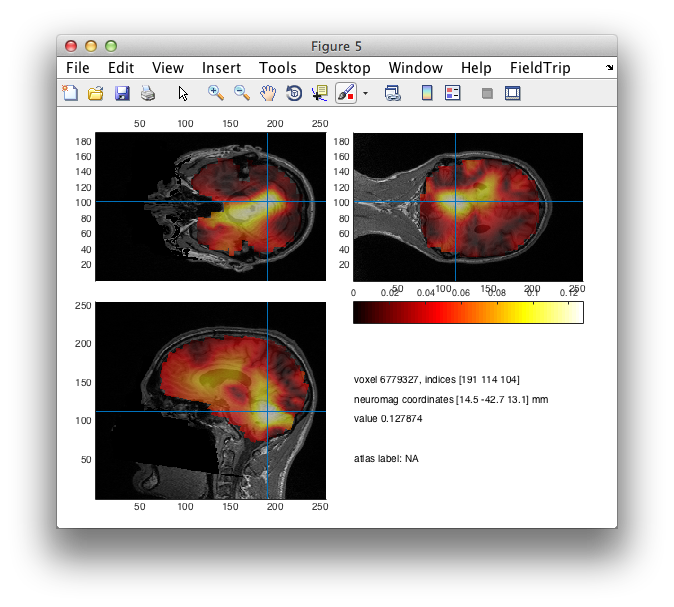
figure; ft_sourceplot(cfg, source_difint);
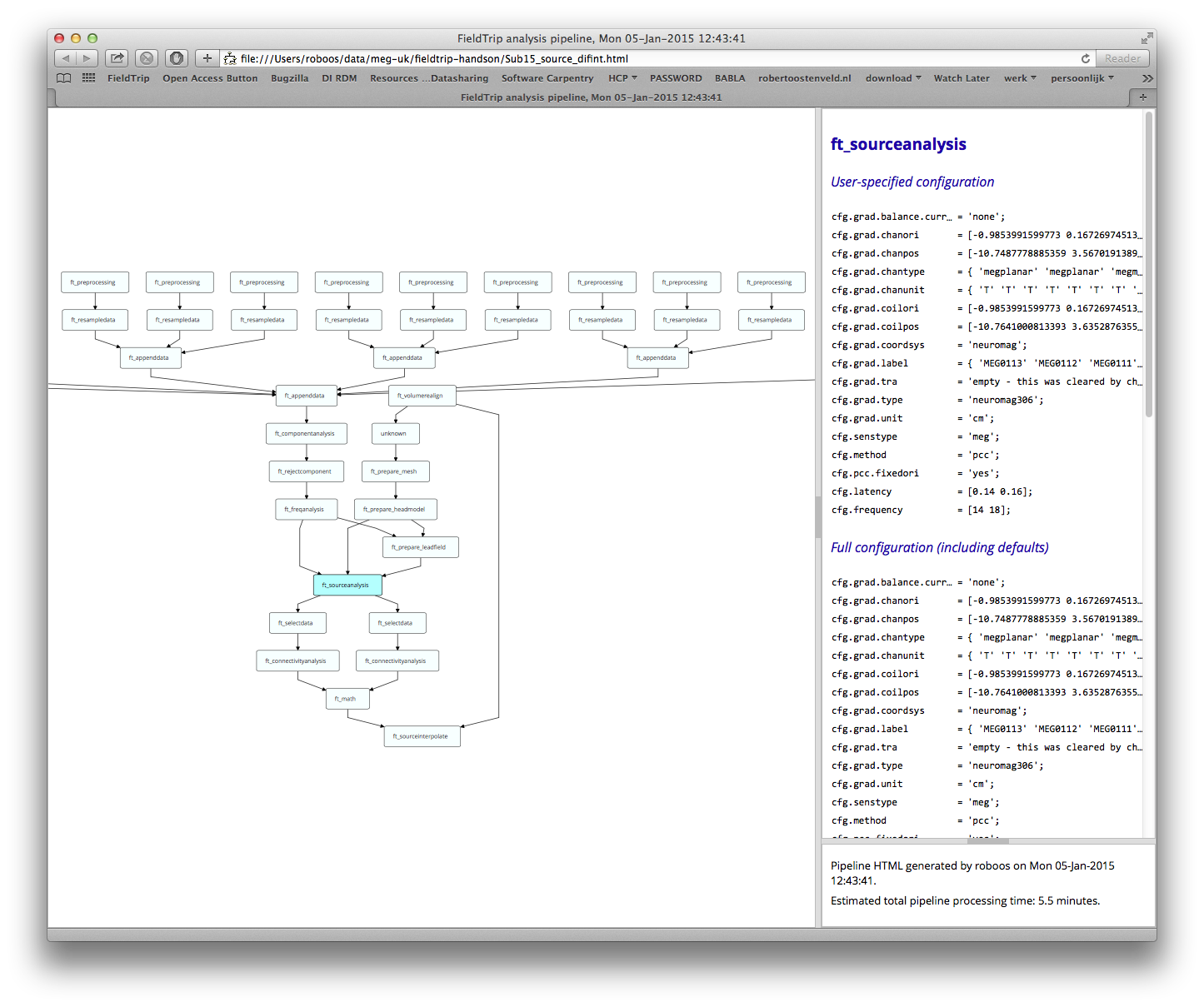
%% look at the analysis history
% for saving to disk
prefix = sprintf('Sub%02d', subj);
cfg = [];
cfg.filename = [prefix '_source_difint.html'];
ft_analysispipeline(cfg, source_difint);