Combine MEG with Eyelink eyetracker data
Description
This example demonstrates how you can combine detailed information from Eyelink eyetracker data with the MEG data to mark eye movement events. The Eyelink eyetracker data contains timing information about blinks, fixations and saccades, which can inform the MEG data analysis, for artifact identification/rejection purposes, or for experimental reasons. Upon data acquisition, the Eyelink can be set up (or is set up by default), to mark saccades and blinks in the output file. This identification scheme is based on certain heuristics (e.g., velocity thresholding of the eye position traces to identify saccades), and may be configured by the researcher. Under the assumption that the parameters have been judiciously specified, it may be useful to use this timing information in the downstream analysis. This could replace an ad-hoc analysis of the eyetracker data that has been recorded along with the MEG data.
Why use information from the Eyelink and not use the eyetracker traces from the MEG data?
The advantage of an ad-hoc analysis of the eyetracker traces that have been collected along with the MEG data, is that the extracted events will be easily synchronized with the timing in the MEG data. This is simply because all data traces have the same shared time axis. The event timings that are stored in the Eyelink file are relative to the Eyelink recording, and need to be mapped onto the timings in the MEG recording. The latter recording might have been started either before or after the Eyelink recording, and moreover will very likely have a different sampling frequency. The example below explains how this mapping can be performed. The advantage of using the marked events from the Eyelink data, is that you don’t need to rely on your own implementation of the artifact marking, which for instance can be achieved using ft_artifact_zvalue on one of the eyetracker signals, in combination with well chosen set of processing parameters. Instead, you will rely on the heuristics that Eyelink has used to mark events.
Procedure
In order to express the timing of the events from the Eyelink file relative to the MEG recording, the following steps are needed:
- Identification of corresponding events in both datasets.
- Computation of the mapping between the two sets of events.
- Adjustment of the Eyelink event times
Example dataset
The dataset used for this example is the first session of the first subject of the Sherlock dataset, which can be downloaded from the Donders Data Repository. The example can be adjusted to use your own data, under the assumption that the converted Eyelink ASC-file (obtained by running EDF2ASC, see the Eyelink getting started page) contains the trigger information sent by the presentation computer during the experiment.
What to do if my experimental data does not contain any triggers, or when the Eyelink data does not contain them?
In this case, you would need to align the signals in a different way. This is not covered by the current example. You could consider to cross-correlate the Eyelink traces with the corresponding traces in the MEG data. This is not trivial at all, due to the difference in sampling frequency.
Identification of corresponding events in both datasets
We start by reading the events from both files.
fname_meg = '/Users/jansch/Desktop/sub-001/ses-001/meg/sub-001_ses-001_task-compr_meg.ds';
fname_asc = '/Users/jansch/Desktop/sub-001/ses-001/eyelink/sub-001_ses-001_eye.asc';
event_meg = ft_read_event(fname_meg);
event_asc = ft_read_event(fname_asc);
The experimental setup at the DCCN is such that triggers, sent by the stimulus presentation computer, end up as events in both datasets. In the MEG data, these are represented as events of the type ‘UPPT001’, and in the Eyelink data these events are known as ‘INPUT’-type events. The Eyelink also explicitly codes the pulse triggers going back to zero as a separate event, so these need to be removed.
selmeg = strcmp({event_meg.type}', 'UPPT001');
event_meg = event_meg(selmeg);
event_asc_all = event_asc;
selasc = strcmp({event_asc.type}', 'INPUT');
event_asc = event_asc(selasc);
val = [event_asc.value];
event_asc = event_asc(val~=0);
Computation of the mapping between the two sets of events
The number of events are not necessarily matched. Under the assumption that a particular subset of the sequence of trigger events is reliably represented in both files, we can investigate whether the event sequences are ‘left’ or ‘right’ aligned. This works only of either one of the recordings has been started early (and may have captured some early triggers not present in the other recording), or one of the recordings has been stopped early (and may not have captured some late triggers, which are present in the other recording). More complicated mapping is not covered here.
val_meg = [event_meg.value];
val_asc = [event_asc.value];
nmin = min(numel(val_meg), numel(val_asc));
if all(val_meg(1:nmin)==val_asc(1:nmin))
% left-aligned
event_meg = event_meg(1:nmin);
event_asc = event_asc(1:nmin);
elseif all(val_meg(end:-1:(end-nmin+1))==val_asc(end:-1:(end-nmin+1)))
% right-aligned
if nmin==numel(event_meg)
event_asc = event_asc((end-nmin+1):end);
event_meg = event_meg(1:nmin);
else
event_meg = event_meg((end-nmin+1):end);
event_asc = event_asc(1:nmin);
end
end
Adjustment of the Eyelink event times
With the trigger events aligned, based on the trigger values, we can estimate the linear mapping between the event times. Assuming that the MEG recording represents a continuous recording, the information in event_meg.sample can be used to unambiguously link the MEG trigger to the MEG data. For the Eyelink data, there is no guarantee that this file is based on a continuous recording (at the time of writing this example, the author has come across some Eyelink with discontinuous time axes, which suggests that the researcher has paused the Eyelink recording during the MEG experiment). For this reason, to play safe, we will use the event_asc.timestamp, rather than event_asc.sample.
smp_asc = [event_asc.timestamp];
smp_meg = [event_meg.sample]; % assuming MEG to be derived from a continuous recording
offset_meg = mean(smp_meg);
offset_asc = mean(smp_asc);
x = smp_asc - offset_asc;
y = smp_meg - offset_meg;
slope = y/x; % this should be about 1.2 (1kHz vs. 1.2kHz)
% plot the residuals between the MEG samples and the 'modelled' MEG samples
res = smp_meg - offset_meg - slope.*(smp_asc - offset_asc); % conclusion: it is anywhere between -1.5 of 1.5 sample
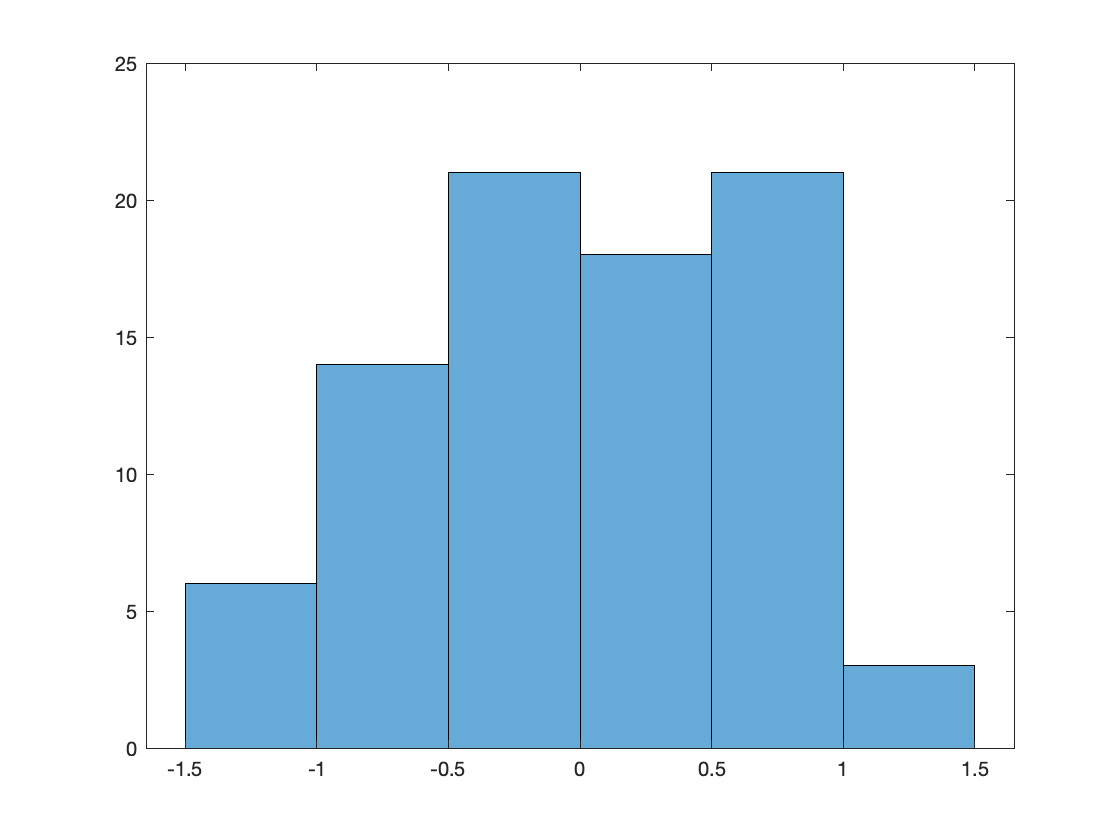
figure;histogram(res);
As can be seen from the histogram, the difference in timing is on the order of less then 1.5 sample (@1.2 kHz), so that looks reasonable.
Note that in this example, the estimated slope is not exactly 1.2, but 1.1999465. This is due to a very small difference in clock speed between the MEG acquisition computer and the Eyelink computer. If the mapping of events would have been based on a value of 1.2, and an alignment of a single early event in both recordings, then the asynchrony further along during the recording can become quite severe, on the order of ~225 ms for a 70 minute recording.
Then, to adjust the timing of all Eyelink events:
S = [event_asc_all.timestamp];
Snew = slope.*(S-offset_asc) + offset_meg;
% remap the event_asc_all's samples to samples of the MEG recording
% adjust the duration from milliseconds to samples, NOTE: this assumes 1kHz
% sampling, i.e. 1 timestamp step is 1 ms.
for k = 1:numel(event_asc_all)
event_asc_all(k).sample = Snew(k);
event_asc_all(k).duration = round(event_asc_all(k).duration.*slope);
end