Creating a layout for plotting NIRS optodes and channels
FieldTrip uses layouts to specify in 2D (e.g., on screen) where channels are to be plotted. This is used for example in ft_multiplotER and ft_multiplotTFR, and also in ft_topoplotER and ft_topoplotTFR. The layout does not only describe where the channel is located (which is required for the topographies), but also how large the channel should be displayed when plotting ERPs on the channel position. Furthermore, it contains an outline to help you to orient (usually a circle indicating the head, and a triangle indicating the nose) and an invisible mask that determines where the data should be topographically interpolated and where not (usually it is the same circle as used for the head outline).
Plotting results of NIRS analysis poses two specific challenges that make it different from EEG and MEG
- there is not a standard layout of the optodes that is always used
- the location of a channel is not so obvious to relate to the optodes
Regarding the first point: in EEG it is common (but not required) to place electrodes according to the 10-20, the 10-10 or the 10-5 system. Most EEG caps are designed to have the electrodes on a subset of the standard locations that usually covers the whole head. Within a lab it is common to use the same cap for all experiments. NIRS systems typically have fewer channels than EEG systems, and whole-head coverage is not so common. Instead, NIRS optodes are usually placed such that they are sensitive to pick up activity from the underlying areas of activity. These areas of activity are study specific, hence different experiments in the same lab would use different placements of the optodes. Consequently, it is required to make a layout for each specific arrangement of optodes.
Regarding the second point: with EEG the electric potential on a channel - although it is a difference between one specific electrode and the reference electrode - is usually plotted on the location of the specific EEG electrode. With MEG the magnetic field is measured by a magnetometer or gradiometer, independent of any reference location. Therefore it is customary to plot the magnetic field on the location of that magnetometer. When the MEG system uses planar gradiometers, as with the Neuromag system, this is not possible since two planar gradiometers are on the same location. In that respect, the situation that we have with NIRS is a bit a combination of both. We have to consider both the transmitter (Tx) and the receiver (Rx) that play a role in the NIRS channel, but also that on one Tx-Rx pair two wavelengths are used, which allows the separation on oxy- and deoxyhemoglobin. Consequently, we have to think about how we want to plot the oxy and deoxy channels in relation to each other. Furthermore, we migth also want to make a distinction in the graphical representation for regular (long) and for short channels.
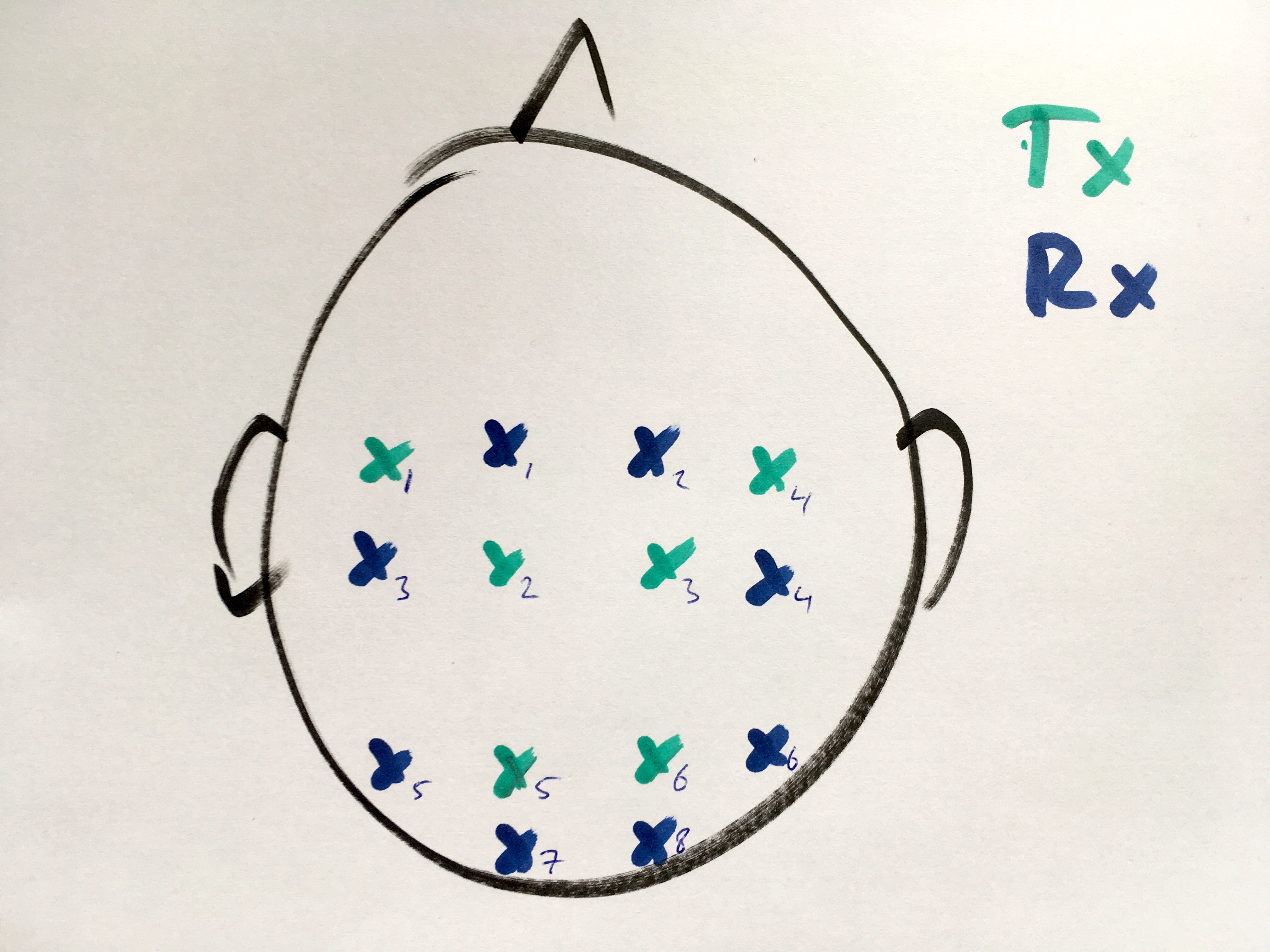
Localize the optodes in a drawing
It is quite easy to make a drawing of the optode positions. You could for example start from an image of the 10-20 system that you can find online.
To localize the optodes in a drawing, we can use the functionality of ft_prepare_layout which was mostly designed for localizing EEG electrodes on a drawing:
cfg = [];
cfg.image = 'optodedrawing.jpg';
layout_optode = ft_prepare_layout(cfg);
- first we use the mouse to click on each of the Tx optodes, then we click on each of the Rx optodes
- press “q” to quit the localization, this brings us to the next step
- click with the mouse on the cornerpoint of each of the polygon contours that we want as (non-visible) mask
- press “c” to close the contour and continue with the next, this starts the localization of the next polygon contour
- press “q” to quit the localization, this brings us to the next step
- click with the mouse on the cornerpoint of each of the contours that we want as (visible) outlines
- press “c” to close the contour (e.g., for the circle) or “n” if we do not want to close it (e.g., with the nose)
- continue making the contours for the head (circle), nose, and for both ears
- after completing the final contour, press “q” to quit
The result looks like this
>> layout
struct with fields:
pos: [16x2 double]
label: {16x1 cell}
width: [16x1 double]
height: [16x1 double]
mask: {[5x2 double] [7x2 double] [0x2 double]}
outline: {[16x2 double] [3x2 double] [6x2 double] [7x2 double]}
cfg: [1x1 struct]
We can use ft_plot_layout to make a figure. You can see that it also added a box for the comment and a box for the scale. These are optional, see ft_prepare_layout.
figure
ft_plot_layout(layout_optode)
This example was made in a bit of a rush, you can imagine that with a little bit more effort you can make it look much nicer.
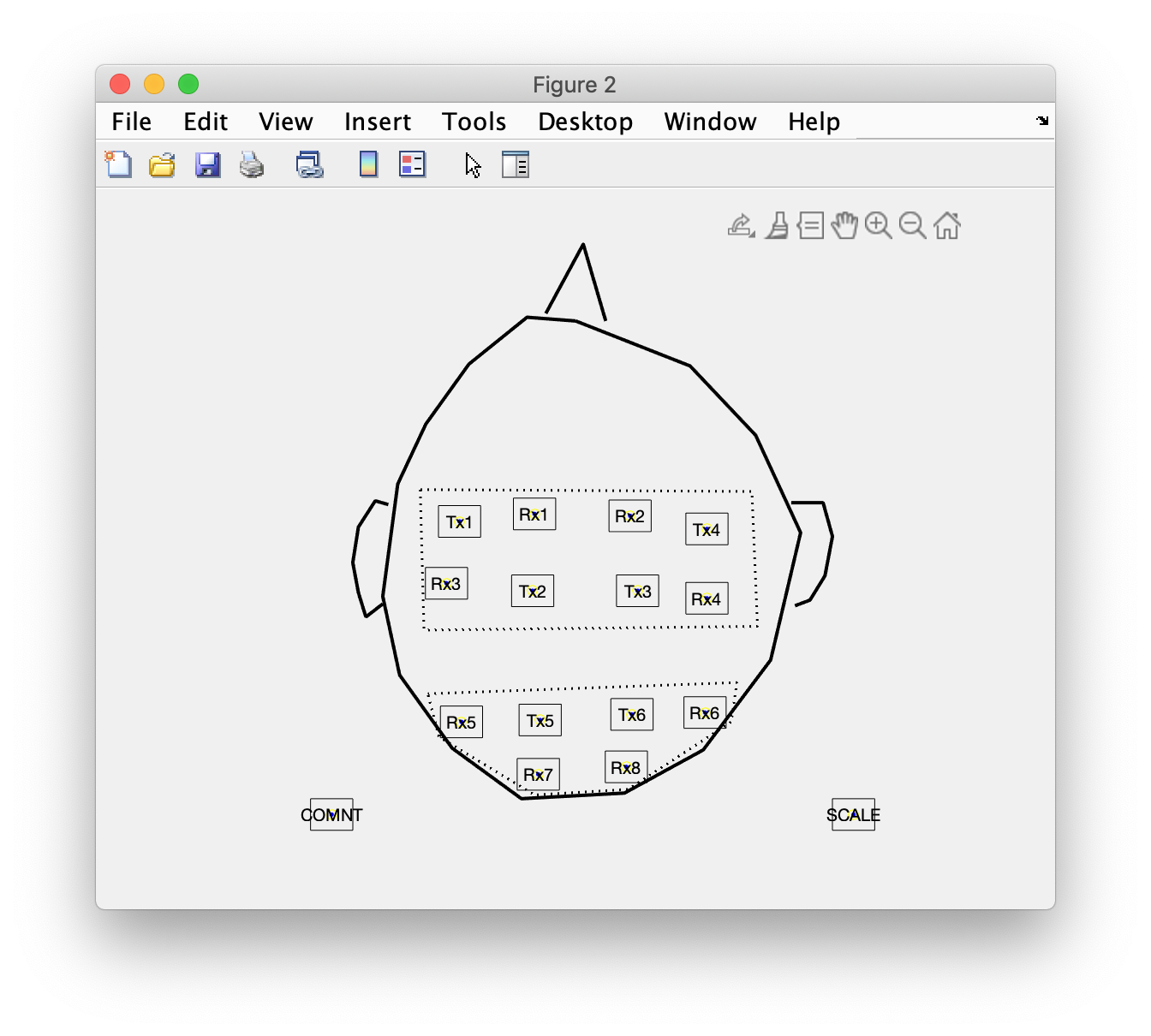
Rename the optode labels
Next we continue with renaming the optode labels.
layout_optode = layout_original;
layout_optode.label(1:6) = {
'Tx1'
'Tx4' % updated, see below
'Tx2' % updated, see below
'Tx3'
'Tx5'
'Tx6'
};
layout_optode.label(7:14) = {
'Rx1'
'Rx2'
'Rx3'
'Rx4'
'Rx5'
'Rx6'
'Rx7'
'Rx8'
};
figure
ft_plot_layout(layout_optode)
This figure initially revealed that I did not click precisely in the order that I had planned. Therefore I updated the list of optode labels, such that it matches the drawing.
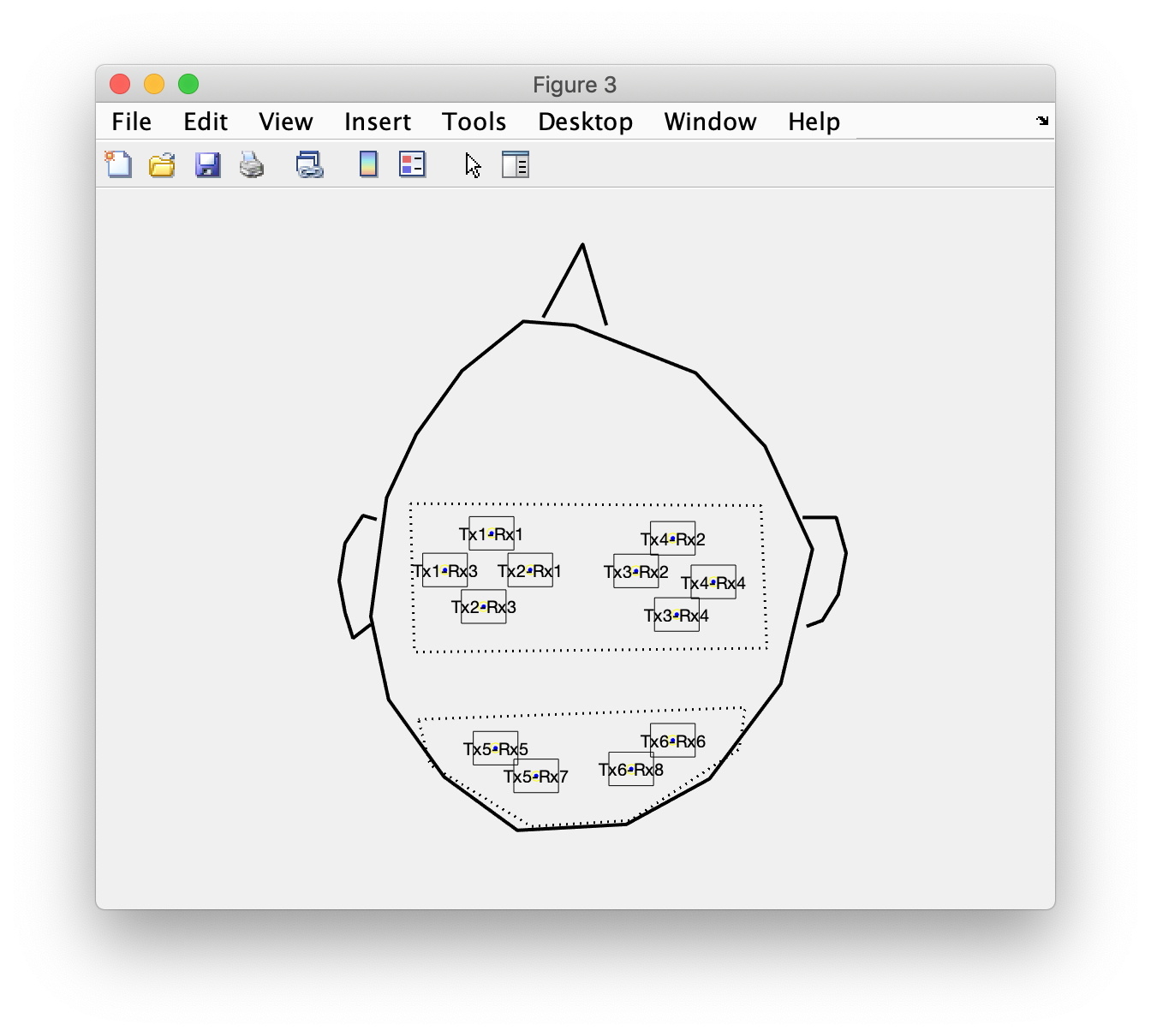
Combine optodes into channels
If this were an EEG layout, we would now be done. However, with NIRS the channels do not correspond directly to the individual optodes, so we need to make combinations of optodes. This can be done using a “montage”, which specifies how optodes (or more commonly EEG electrodes) are combined in a channel. The concent of a montage is explained in more detail in ft_apply_montage.
montage.labelold = {'Tx1', 'Tx2', 'Tx3', 'Tx4', 'Tx5', 'Tx6', 'Rx1', 'Rx2', 'Rx3', 'Rx4', 'Rx5', 'Rx6', 'Rx7', 'Rx8'};
Making the montage is quite a puzzle and probably requires some trial and error. Each row corresponds to a (new) channel, each column to an (old) optode. With +1 and -1 you can indicate how the channels are weighted; for EEG or iEEG this would correspond to making bipolar derivations.
montage.tra = [
%Tx1 Tx2 Tx3 Tx4 Tx5 Tx6 Rx1 Rx2 Rx3 Rx4 Rx5 Rx6 Rx7 Rx8
1 0 0 0 0 0 -1 0 0 0 0 0 0 0
1 0 0 0 0 0 0 0 -1 0 0 0 0 0
0 1 0 0 0 0 -1 0 0 0 0 0 0 0 % Tx2 goes to Rx1
0 1 0 0 0 0 0 0 -1 0 0 0 0 0 % Tx2 goes to Rx3
0 0 1 0 0 0 0 -1 0 0 0 0 0 0
0 0 1 0 0 0 0 0 0 -1 0 0 0 0
0 0 0 1 0 0 0 -1 0 0 0 0 0 0
0 0 0 1 0 0 0 0 0 -1 0 0 0 0
0 0 0 0 1 0 0 0 0 0 -1 0 0 0
0 0 0 0 1 0 0 0 0 0 0 0 -1 0
0 0 0 0 0 1 0 0 0 0 0 -1 0 0
0 0 0 0 0 1 0 0 0 0 0 0 0 -1
];
The new channel labels can be deduced from the +1 and -1 occurences in each row:
montage.labelnew = cell(size(montage.tra,1), 1);
for i=1:size(montage.tra,1)
label_plus = montage.labelold{montage.tra(i,:)== 1};
label_minus = montage.labelold{montage.tra(i,:)==-1};
montage.labelnew{i} = [label_plus '-' label_minus];
end
Using this montage, we can construct the layout with the channels halfway between the transmitter and receiver optodes.
cfg = [];
cfg.layout = layout_optode;
cfg.montage = montage;
cfg.skipscale ='yes'; % this will be added later again
cfg.skipcomnt ='yes'; % this will be added later again
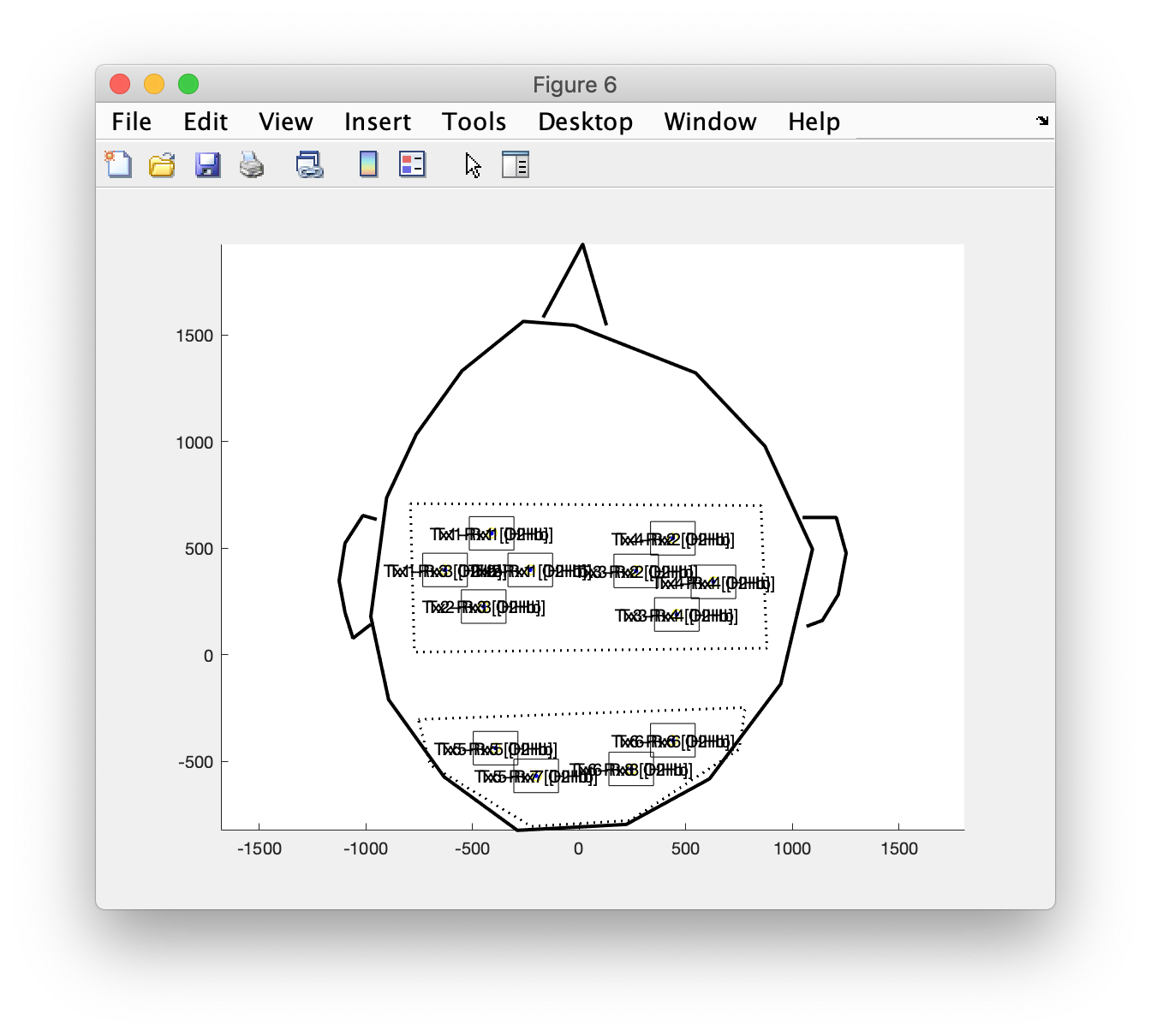
layout_channel = ft_prepare_layout(cfg);
Again you can plot the layout, probably you will have to do this repeatedly while updating the montage.tra matrix above
figure
ft_plot_layout(layout_channel)
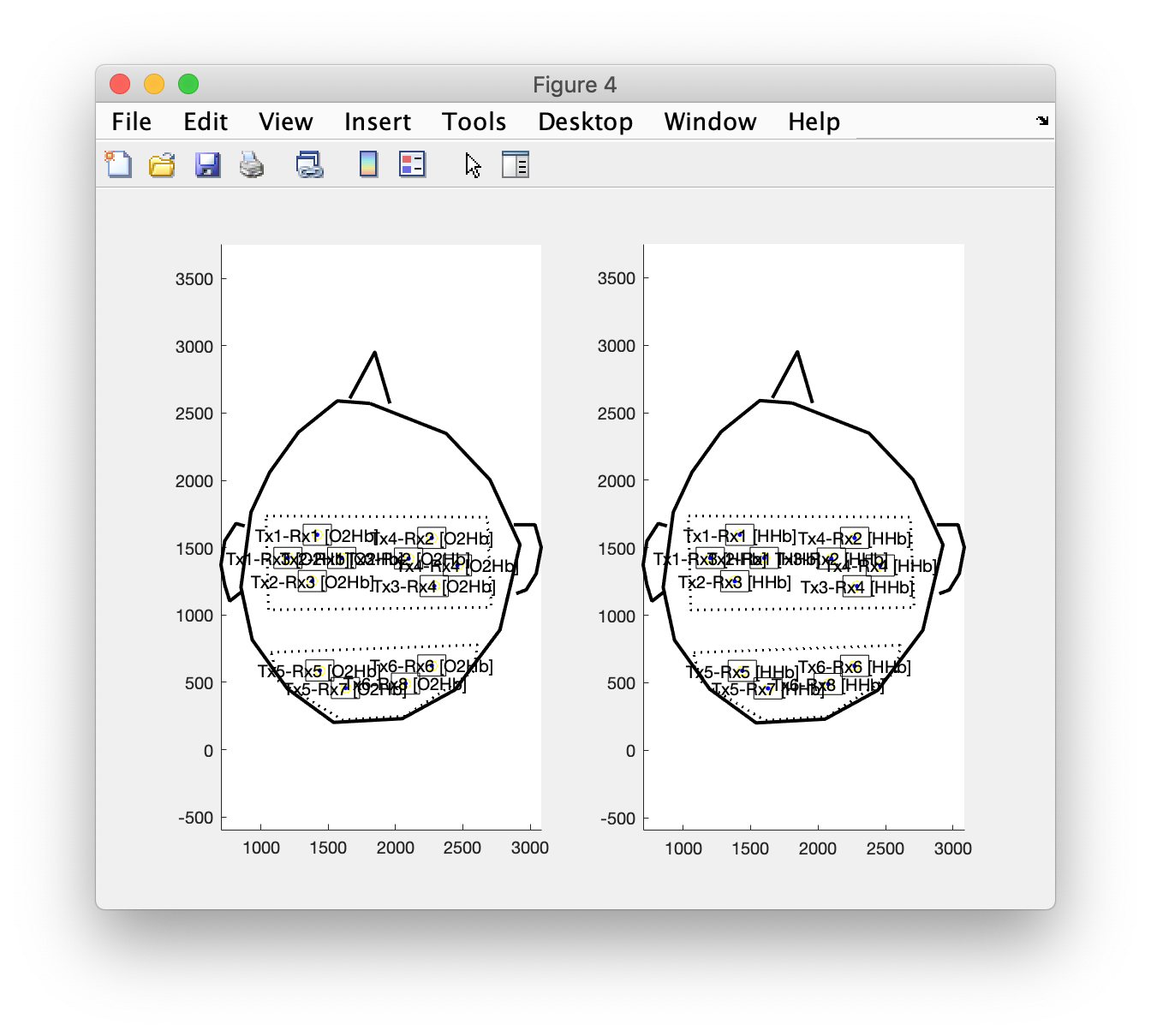
Make separate channels for the oxy- and deoxyhemoglobin (or wavelengths)
We are still not ready, since we need to add separate channels for the optical densities (OD) for the two wavelengths, or after ft_nirs_transform_ODs for the oxyhemoglobin (O2Hb) and deoxyhemoglobin (HHb).
figure
layout_channel_O2Hb = layout_channel;
for i=1:numel(layout_channel_O2Hb.label)
layout_channel_O2Hb.label{i} = [layout_channel_O2Hb.label{i} ' [O2Hb]']; % or with the wavelength
end
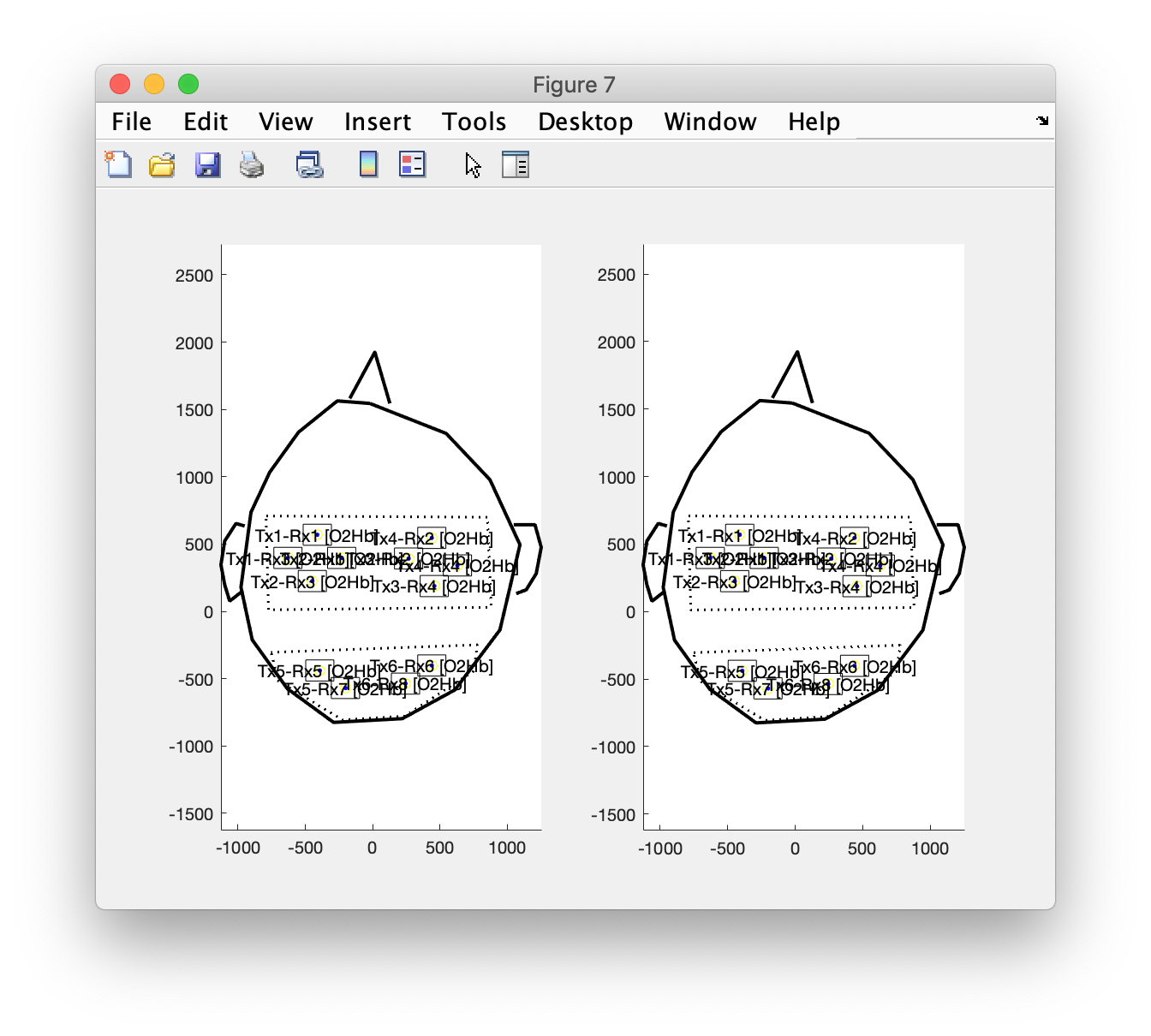
subplot(1,2,1)
ft_plot_layout(layout_channel_O2Hb)
axis on
layout_channel_HHb = layout_channel;
for i=1:numel(layout_channel_HHb.label)
layout_channel_HHb.label{i} = [layout_channel_HHb.label{i} ' [HHb]']; % or with the wavelength
end
subplot(1,2,2)
ft_plot_layout(layout_channel_HHb)
axis on
Combine the oxy- and deoxyhemoglobin layouts
We can combine the two layouts in a single layout with ft_appendlayout.
cfg = [];
cfg.direction = 'horizontal'; % or vertical
cfg.distance = 1000; % this required some trial and error, it is expressed in pixels of the original image
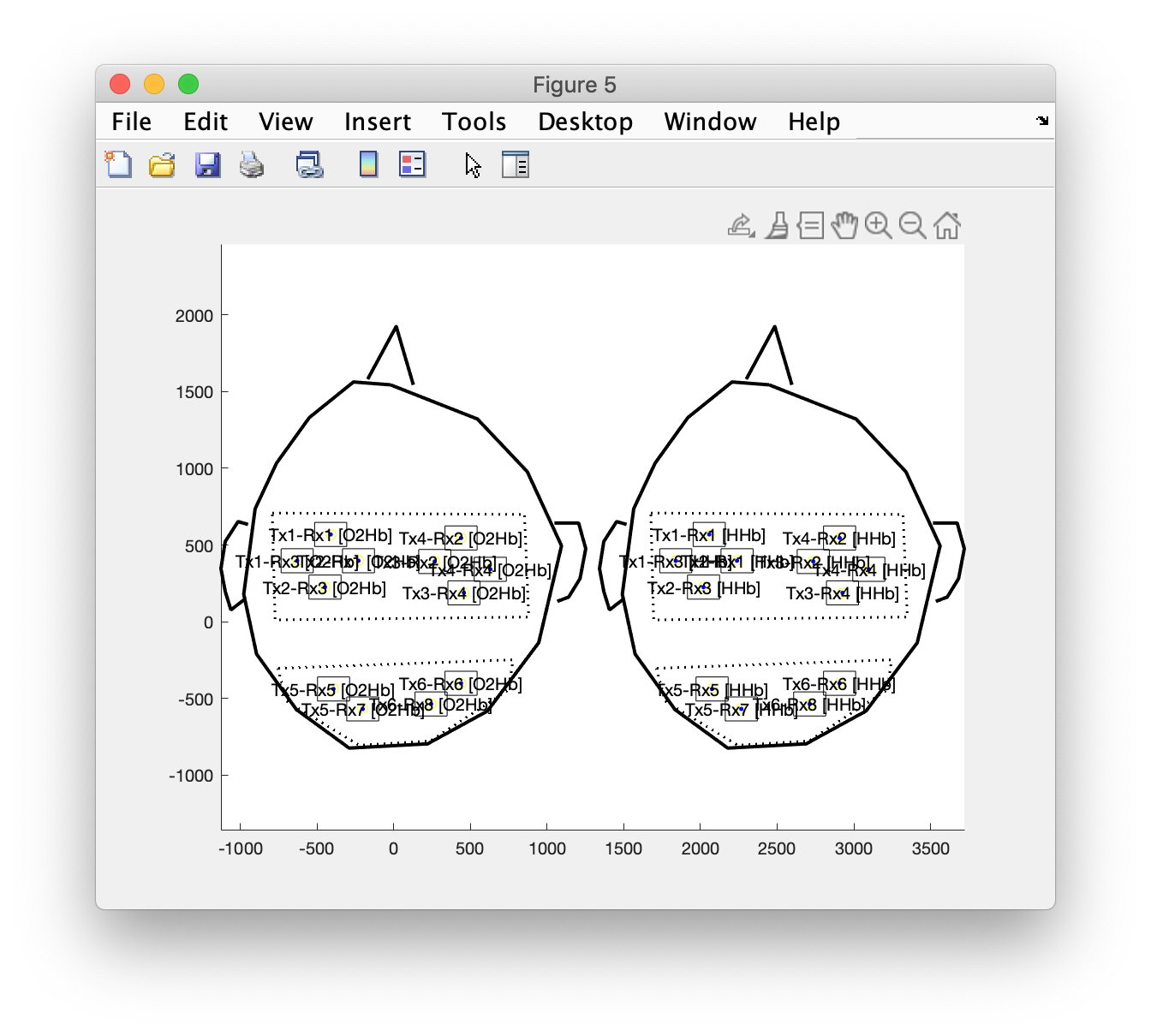
layout_channel_v1 = ft_appendlayout(cfg, layout_channel_O2Hb, layout_channel_HHb);
figure
ft_plot_layout(layout_channel_v1);
axis on % this makes it explicit that it is now in a single axis
It is also possible to plot the oxy and deoxy channel on top of each other, e.g. using different line colors. For that you have to specify a distance of 0.
cfg = [];
cfg.direction = 'overlapping';
layout_channel_v2 = ft_appendlayout(cfg, layout_channel_O2Hb, layout_channel_HHb);
figure
ft_plot_layout(layout_channel_v2);
axis on
The channels are now at exactly the same position and the labels appear fuzzy because they are printed on top of each other.
The same procedure can be used to make the layouts for the two different infrared wavelengths, and these can be combined side-by-side or overlapping as well.
Split the oxy- and deoxyhemoglobin into separate layouts
If you have a layout that already combines the oxy and deoxy channels in a particular way (e.g., overlapping) and you want to change it (e.g., into side-by-side), you can use the following code to split the different channels again.
layout_channel_O2Hb = layout_channel_v2; % start with a copy of the combined one
sel = contains(layout_channel_O2Hb.label, 'O2Hb');
layout_channel_O2Hb.label = layout_channel_O2Hb.label (sel);
layout_channel_O2Hb.pos = layout_channel_O2Hb.pos (sel,:); % this is an Nx2 array
layout_channel_O2Hb.width = layout_channel_O2Hb.width (sel);
layout_channel_O2Hb.height = layout_channel_O2Hb.height(sel);
layout_channel_HHb = layout_channel_v2; % start with a copy of the combined one
sel = contains(layout_channel_HHb.label, 'O2Hb');
layout_channel_HHb.label = layout_channel_HHb.label (sel);
layout_channel_HHb.pos = layout_channel_HHb.pos (sel,:); % this is an Nx2 array
layout_channel_HHb.width = layout_channel_HHb.width (sel);
layout_channel_HHb.height = layout_channel_HHb.height(sel);
figure
subplot(1,2,1)
ft_plot_layout(layout_channel_O2Hb)
axis on
subplot(1,2,2)
ft_plot_layout(layout_channel_HHb)
axis on
Now we are back at the situation we were before with two separate layouts, and you can again use ft_appendlayout (see above) to combine the two layouts either overlapping or side-by-side.
Making a layout from 3D optode positions]
If you are starting with a structure sensor recording of the 3D optode positions as demonstrated in the tutorial on localizing electrodes using a 3D-scanner, then you can start by making a layout for only the Tx and Rx optodes, From that you can subsequently construct the channels (see above) from the pairwise optode combinations, and again construct the layout for the oxy- and deoxyhemoglobin.
Summary and further reading
This example demonstrated how to make a layout for plotting NIRS data, starting from a drawing. It also shows how you can take an existing layout, split it between the channel types and modify it.
Here are some other tutorials that provide useful information: