Should I rereference prior to or after ICA for artifact removal?
The short answer is: It does not really matter. There are however a few things to keep in mind.
Let’s assume your data was recorded with a system comprising N channels with the Nth channel being the reference. From this, one can derive N-1 unique time series:
X_i(t) = channel_i(t) - channel_N(t) and i = 1, … , N-1.
Performing an ICA on this data will maximally yield N-1 unique independent components. In fact, it is by definition not possible to derive more independent components than there are linearly independent signals. In other words, the number of independent components one can compute is bounded by the rank of the data. This has implications for when we rereference the data to the average reference before computing the ICA:
Xre_i(t) = X_i(t) - 1/(N-1) sum_j^(N-1) X_j
The vectors Xre_i are linearly dependent since sum_i^(N-1) Xre_i = 0. By rereferencing to the average reference, the rank of the data is reduced by one and therefore, only N-2 independent components could be derived. There is a trick, however, that allows one to obtain N-1 components even for the rereferenced data: Adding a row of zeros to the original (i.e. non-rereferenced) data does neither change its rank nor does this effectively add artifactual information. What does change, however, is the fact that rereferencing to the new average (i.e., including the line of zeros) will not reduce the rank of the data by one.
ICA is often used as a means for artifact removal. In this case, certain independent components are removed and the remaining ones are projected back to the original electrode space. One could now pose the question, whether the independent components one obtains from the ICA differ depending on whether the input data was rereferenced or not. It turns out that due to the nature of ICA-algorithms, the ICs obtained in the two cases will in general be very similar but not identical. It is safe to assume that obvious artifactual (and other meaningful) components will be detected and can thus be removed (or kept) in both cases. Therefore, rereferencing to the average reference after ICA for artifact removal will lead to very similar result as compared to not rereferencing before the ICA.
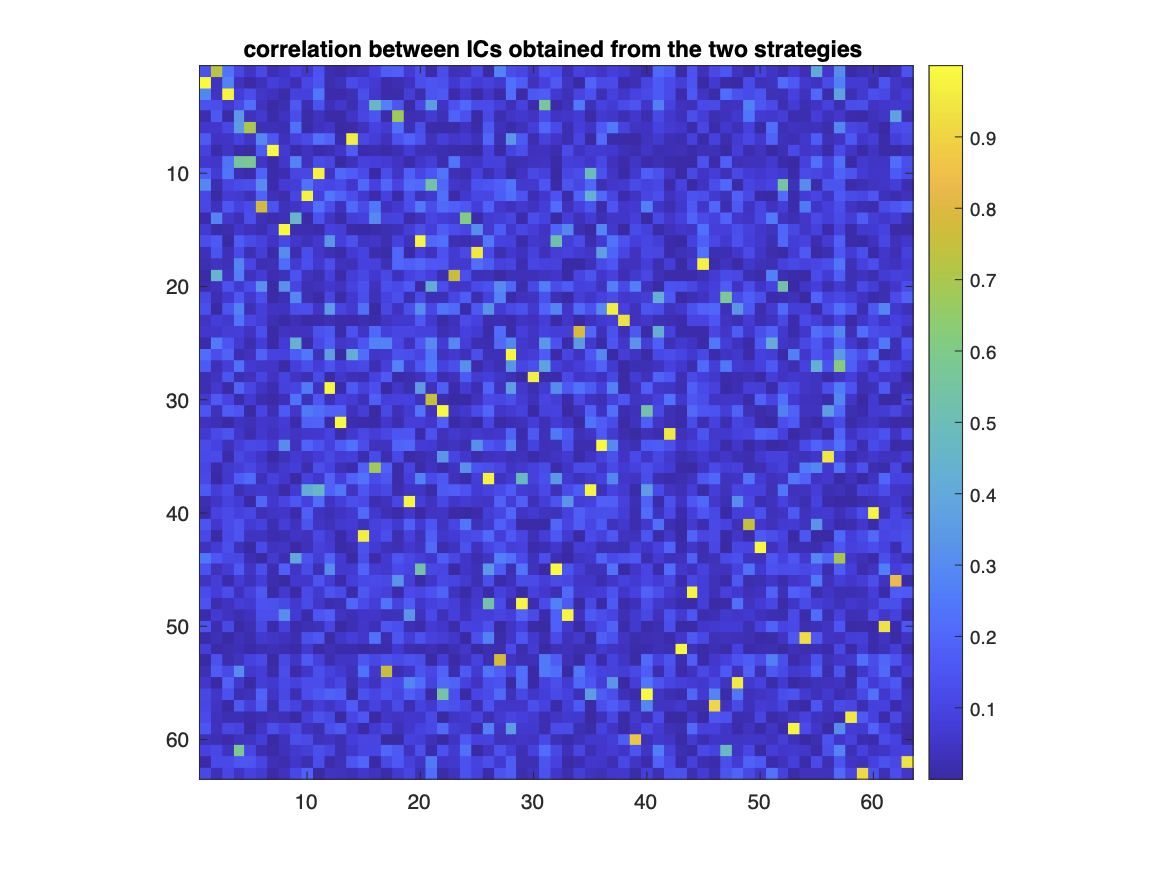
In the following, an illustration of the above, theoretical explanation is provided. First, we have computed the correlation between independent components obtained from rereferenced and non-rereferenced data (Fig 1).
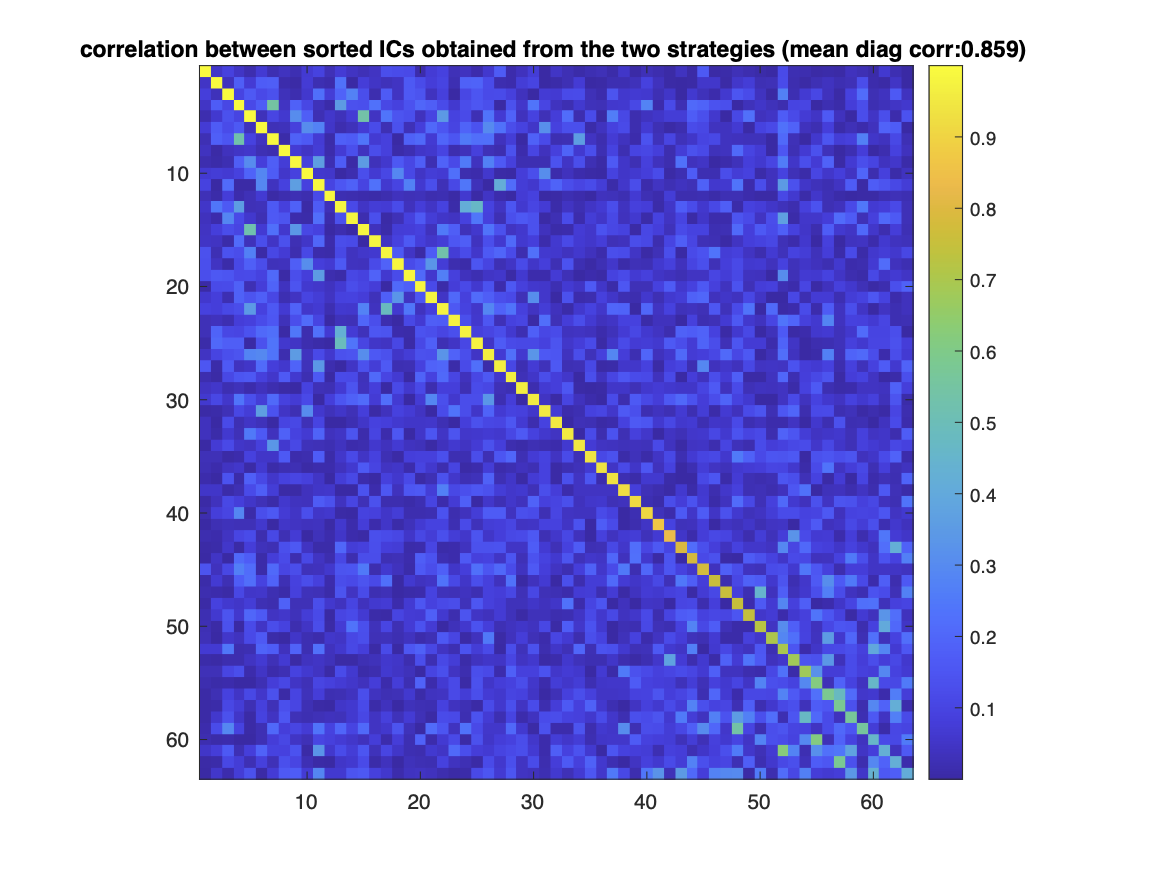
It can be seen that in each line of the correlation matrix there is one correlation value that is significantly larger than the rest. This means that each element in one set of ICs has a matching counterpart in the other set. This can be made clearer by rearranging the lines and columns of the correlation matrix such that 1.) the index of a certain component in one set matches the index of its counterpart in the other set and 2.) the indices are ordered according to the descending order of correlation values (Fig. 2).
The dominant diagonal illustrates that the ICA is relatively invariant with respect to the rereferencing.
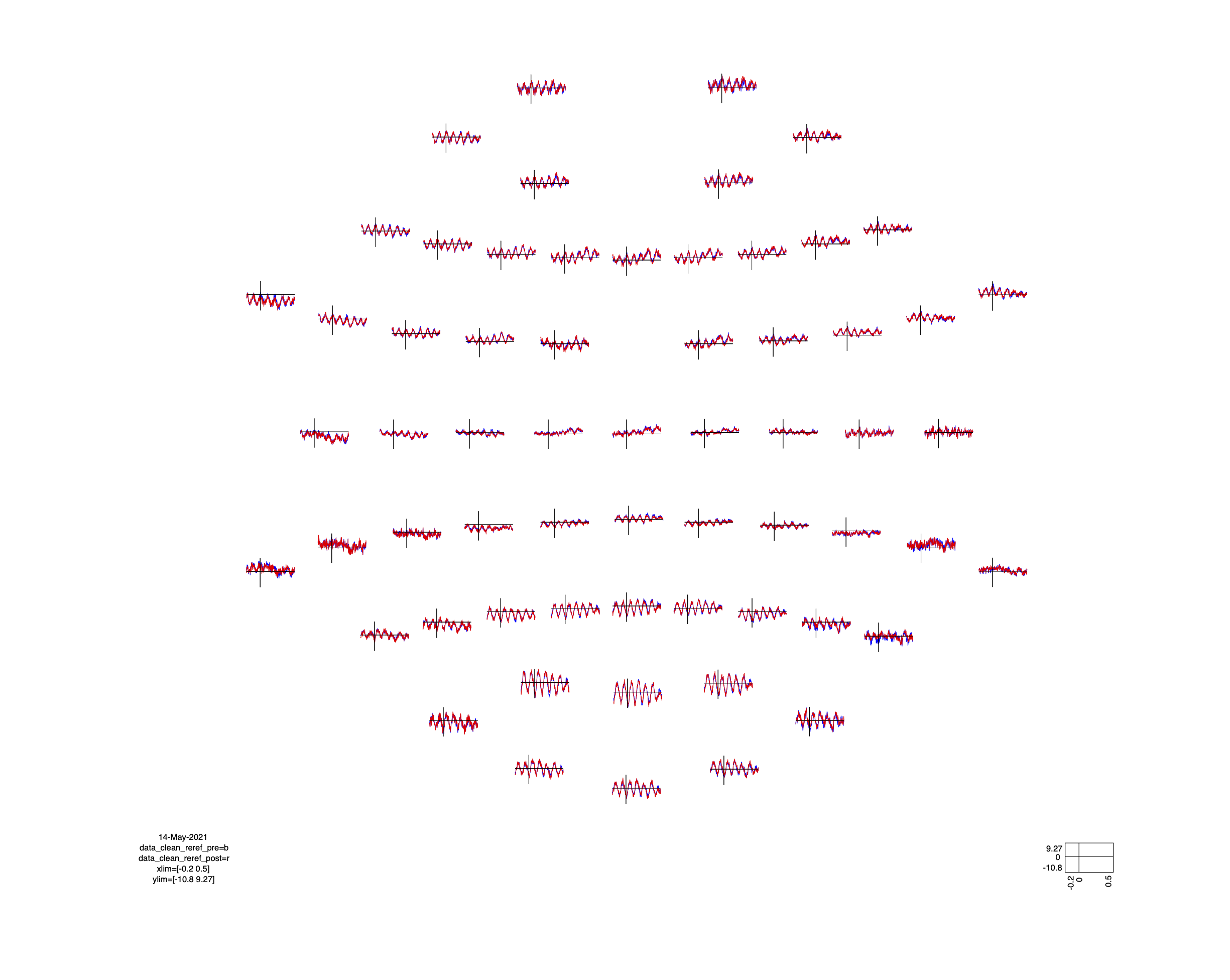
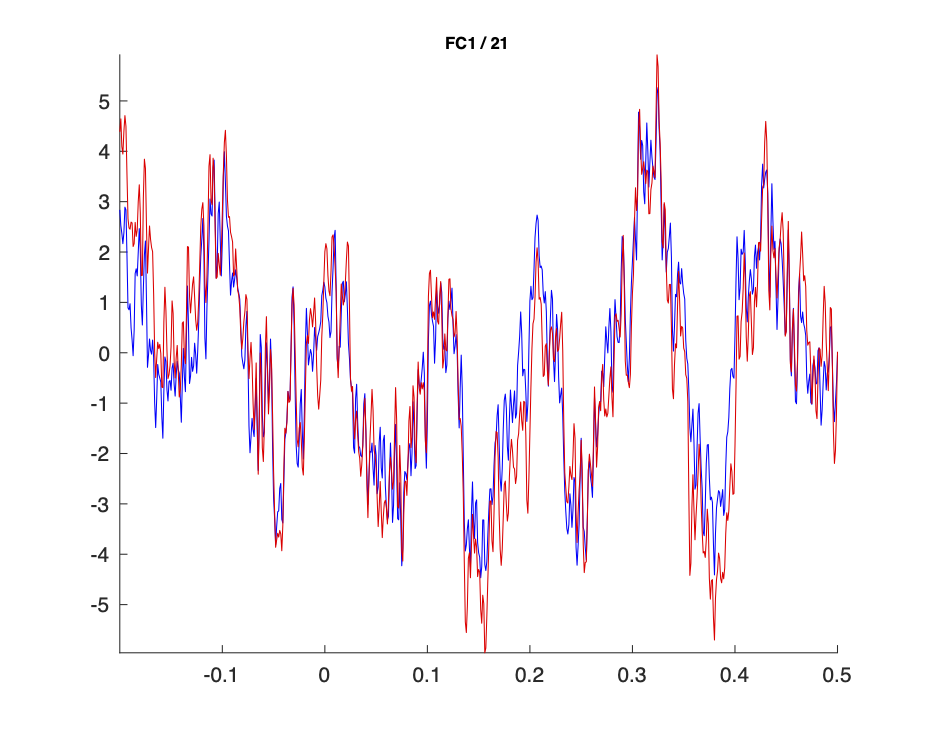
In a next step, we randomly selected roughly half of the matching pairs from both sets of ICs, removed those and computed the inverse ICA. In Fig. 3 and Fig. 4 it can be seen, again, that the results obtained from the two strategies (i.e. the “clean data”) are close to identical.
Code
In the following, the code used to generate above figures is provided. The example data used in this example can be downloaded here.
load data_selectedTrial % load data
load ../fieldtrip/template/layout/acticap-64ch-standard2.mat % load electrode layout
% make sure results are reproducible
seed = 1;
rng(seed);
% determine number of channels
nChannel = length(data_selectedTrial.label);
% add row of zeros to the data
cfg = [];
cfg.implicitref = 'my_implicitref';
data_selectedTrial = ft_preprocessing(cfg, data_selectedTrial);
% rereference individual trials to average reference
cfg = [];
cfg.channel = 'all'; % this is the default
cfg.reref = 'yes';
cfg.refmethod = 'avg';
cfg.refchannel = 'all';
data_selectedTrial_avgreref = ft_preprocessing(cfg, data_selectedTrial);
% compute ICAs ============================================================
cfg = [];
cfg.method = 'runica';
cfg.numcomponent = nChannel;
comp_selectedTrial = ft_componentanalysis(cfg, data_selectedTrial);
comp_selectedTrial_avgreref = ft_componentanalysis(cfg, data_selectedTrial_avgreref);
% visulaize ICs ===========================================================
% compute correlation coefficient between all component time courses
correlation = corr(comp_selectedTrial.trial{1}', comp_selectedTrial_avgreref.trial{1}');
correlation_abs = abs(correlation);
% determine maximum correlation values in each column of the correlation
% matrix
[max_vals, max_inds] = max(correlation_abs);
% Order according to degree of correlation
[sortedVal, sortedInd] = sort(diag(correlation_abs(max_inds, :)), 'descend');
mean_diag_corr = num2str(mean(diag(correlation_abs(max_inds(sortedInd), sortedInd))),3);
figure;
imagesc(correlation_abs);
axis square
colorbar;
pause(0.5) % for some weird reason omitting this lead to the title not being displayed
title(['correlation between ICs obtained from the two strategies'])
% plot
figure;
imagesc(correlation_abs(max_inds(sortedInd), sortedInd));
axis square
colorbar;
pause(0.5) % for some weird reason this is required to show an image title
title(['correlation between sorted ICs obtained from the two strategies (mean diag corr:', mean_diag_corr, ')'])
% reject ICs ==============================================================
nRemove = 30;
remove_inds = randperm(length(comp_selectedTrial.label), nRemove);
% reject components in comp_selectedTrial
cfg = [];
cfg.component = max_inds(sortedInd(remove_inds)); % to be removed component(s)
data_clean = ft_rejectcomponent(cfg, comp_selectedTrial, data_selectedTrial);
% rereference trials in data_clean to average reference
cfg = [];
cfg.channel = 'all'; % this is the default
cfg.reref = 'yes';
cfg.refmethod = 'avg';
cfg.refchannel = 'all';
data_clean_reref_post = ft_preprocessing(cfg, data_clean);
% reject components in comp_selectedTrial_avgreref
cfg = [];
cfg.component = sortedInd(remove_inds); % to be removed component(s)
data_clean_reref_pre = ft_rejectcomponent(cfg, comp_selectedTrial_avgreref, data_selectedTrial_avgreref);
% Visulaize results ===================================================
cfg = [];
cfg.layout = lay;
cfg.xlim = [-0.2, 0.5];
ft_multiplotER(cfg, data_clean_reref_pre, data_clean_reref_post)