development / project / parallel /
Improve parallel computing under the hood
The purpose of this page is just to serve as todo or scratch pad for the development project and to list and share some ideas.
After making changes to the code and/or documentation, this page should remain on the website as a reminder of what was done and how it was done. However, there is no guarantee that this page is updated in the end to reflect the final state of the project
So chances are that this page is considerably outdated and irrelevant. The notes here might not reflect the current state of the code, and you should not use this as serious documentation.
For EEG and MEG research we often process large amount of data, over multiple subjects, and using different analysis strategies implemented in pipelines. Executing such a pipeline, or reexecuting it completely once all parameters have been fixed, is a tedious task due to the computational costs.
The currently advertised strategy to speed up large computations is to parallelize over subjects, i.e. at the coarsest level of granularity. The disadvantage is that it requires quite some coding skills from researchers: the analysis scripts have to be written in a smart fasion to be able to execute them both on a single computer (for development) and on a big and/or distributed compute system for the complete analysis. If distributed or parallel computing were possible under the hood of the FieldTrip functions, many more people would be able to use it, without the necessity to modify the structure of their analysis scripts.
Organization of the end-user’s analysis pipelines
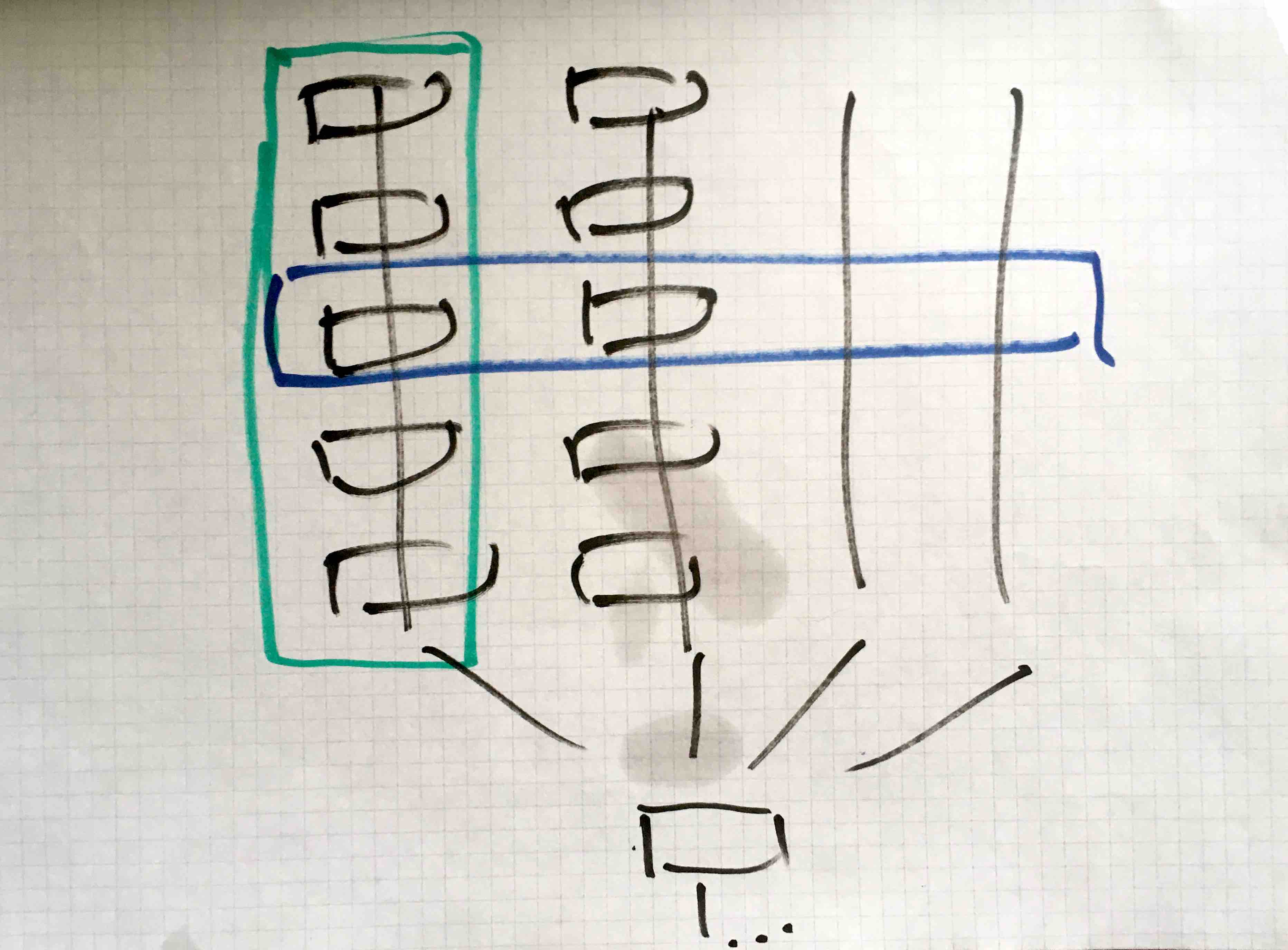
Most EEG/MEG projects include the analysis of data from multiple subjects, followed by an analysis at the group level. If each subject analysis for example consists of 5 steps, you can conceptualize the combination of all analysis as follows.
The researcher can first analyze one subject completely (green box), and then move on with the next. Or the researcher can do a certain step for all subjects (blue box), and then move on with the next step for all subjects.
In my (=Robert’s) experience, the researchers will switch between the two strategies during the development of their analysis pipeline.
Organization of the analysis toolbox
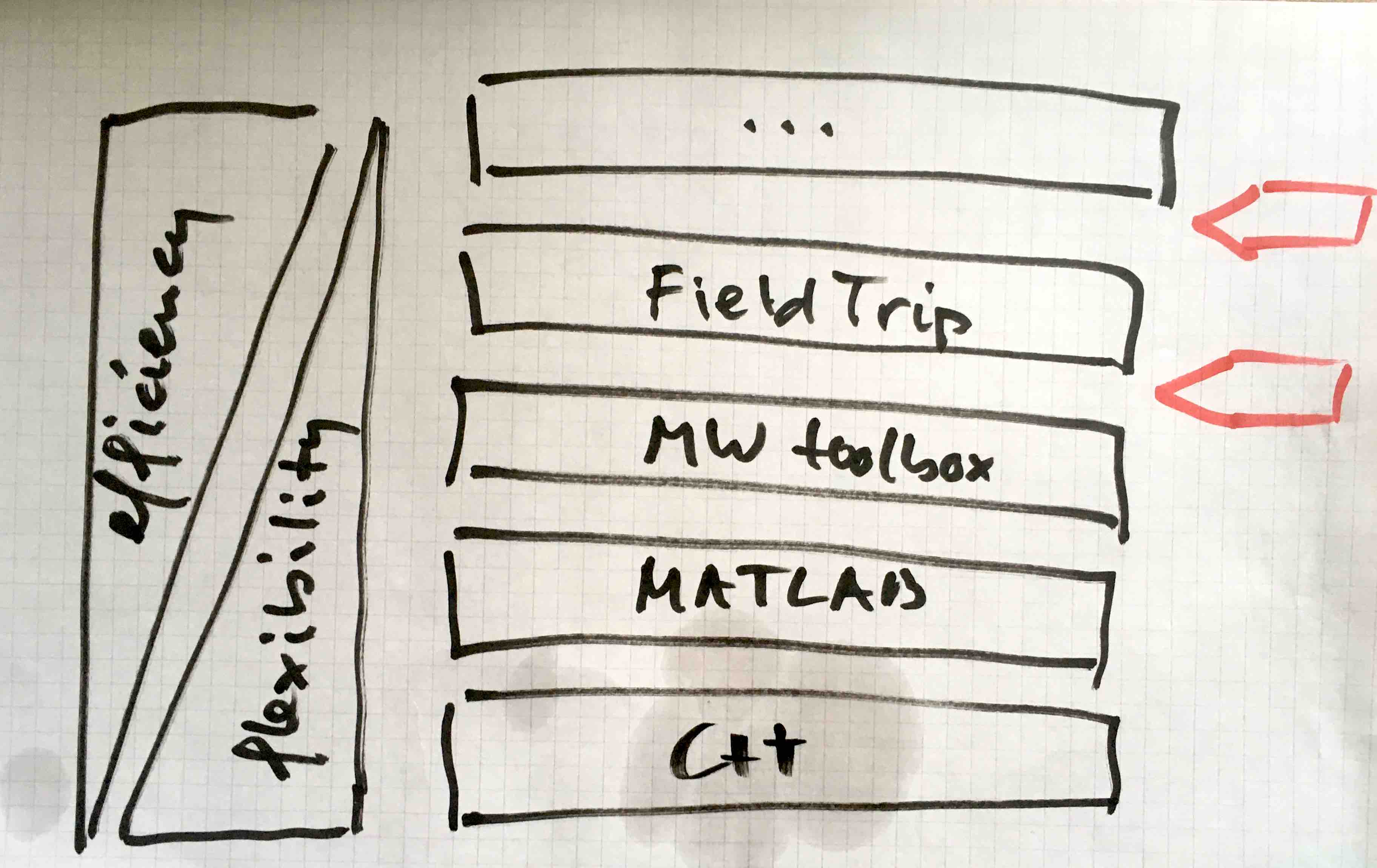
The user writes a script that is one level above the analysis toolboxes like FieldTrip, i.e. the user’s script is at the top. The code in FieldTrip is stacked onto MathWorks toolboxes, which is stacked onto the core MATLAB language, which is stacked onto C++, etc.
The efficiency (speed for the user to implement something) increases when going up, flexibility for the user increases when going down. Power-users are comfortable at the lower levels, whereas novice users prefer clicking in a graphical user interface at the top.
The parallelization can be implemented on the level just above the analysis toolbox (i.e. FieldTrip), or at the level just below. Right now we assume that FieldTrip users parallelize in their own analysis scripts, i.e. above the toolbox layer. This means that the user needs to incorporate the complexity of distributed computing in his/her code.
Distributing computations over subjects
If a researcher has a single do-single-subject-analysis script, it is not too hard to start multiple instances of that script on multiple computers, or to schedule them as a batch on a compute cluster. In this case there is some bookeeping at the level of the batch (start a job for each subject). Each parallel job does its own data bookkeeping, usually this involves writing and reading data to/from disk.
Parallel computing in each high-level function
Rather than doing the parallelization at the level of the users’ scripts (the top red arrow in the figure above), an alternative is to parallelize closer to the actual implementation of the algorithm just below the FieldTrip function interface (the bottom red arrow in the figure above). In this case it would be below the level of bookkeeping by the user in his/her script, and preferably also just below the level of the (shared) bookeeping with regard to the data structure handling. The batch here would e.g., consist of doing a FFT for every trial, computing some feature for each channel, or scanning a section of the brain (rather than the whole brain).
Possibly this could be implemented something like this:
poolobj = parpool;
cfg = [];
cfg.method = 'mtmfft';
cfg.taper = 'hanning';
cfg.foilim = [0 100];
cfg.trials = 1:100;
cfg.parallel = poolobj; % this allows looking up the pool details and number of workers
freq = ft_freqanalysis(cfg, data)
delete(poolobj);
This would distribute 100 jobs for the 100 trials over the workers in the parallel pool.
Setting up the parallel pool would be done using standard MATLAB commands. It would also be possible for the researcher to mix and match the cfg.parallel option in the FieldTrip functions with their own use of parfeval or parfor.
To be done
- identify which high-level functions take most time, e.g., by profiling the tutorial code
- identify which high-level functions have a similar structure (so that the code refactoring can be reused)
- set up an environment that allows for parallel execution on a single multi-core computer
- set up an environment that allows for parallel execution on a typical compute cluster
- implement parallelization in some functions
- evaluate the performance gain
- implement parallelization in more functions
- update the distributed computing guidelines on the website
Results
The results of this project are organized and discussed on the following places:
- GitHub project 5
- GitHub issue 1851, 1852, 1853, 2068, 2069
- the repositories https://github.com/apjanke/fieldtrip-parallel-support and https://github.com/AljenU/fieldtrip-parallel-support
Following the evaluation of the parfor implementation in a number of high-level FieldTrip functions, we decided at this moment not to merge this in the release branch and not continue with a more wide-spread implementation. The impact on the flow of the code is too significant, and interferes with efficient memory handling. The implementations that have been made are maintained for posterity in a separate parallel branch.