Using reproducescript on a full study
This example script will introduce you to functionality in the FieldTrip toolbox designed to aid in making your analysis pipeline - including code, data and results - easily reproducible and shareable. It is based on the manuscript Reducing the efforts to create reproducible analysis code with FieldTrip. We assume that you already had a look at the examples on Making your analysis pipeline reproducible using reproducescript and Using reproducescript for a group analysis.
Example 3
Example 1 and 2 were fairly simple analysis pipelines that didn’t particularly benefit from reproducescript because the original scripts and data organisation were already clear. Those examples are intended to show how reproducescript works and how it’s used. In this example, we apply reproducescript to a published analysis pipeline of MEG data by Andersen (2018). We hope that this will help you to set up your own analysis pipeline using reproducescript.
Original analysis
The analysis pipeline in Andersen (2018) is well-documented and itself a good demonstration of a reproducible analysis pipeline in the FieldTrip ecosystem. Nevertheless, it consists of a complex set of 10 analysis scripts and 46 functions, which, without the extensive documentation that has been provided by the author, would be challenging to reuse and reproduce the results. This makes it particularly suited to demonstrate the effectiveness and simplicity of reproducescript.
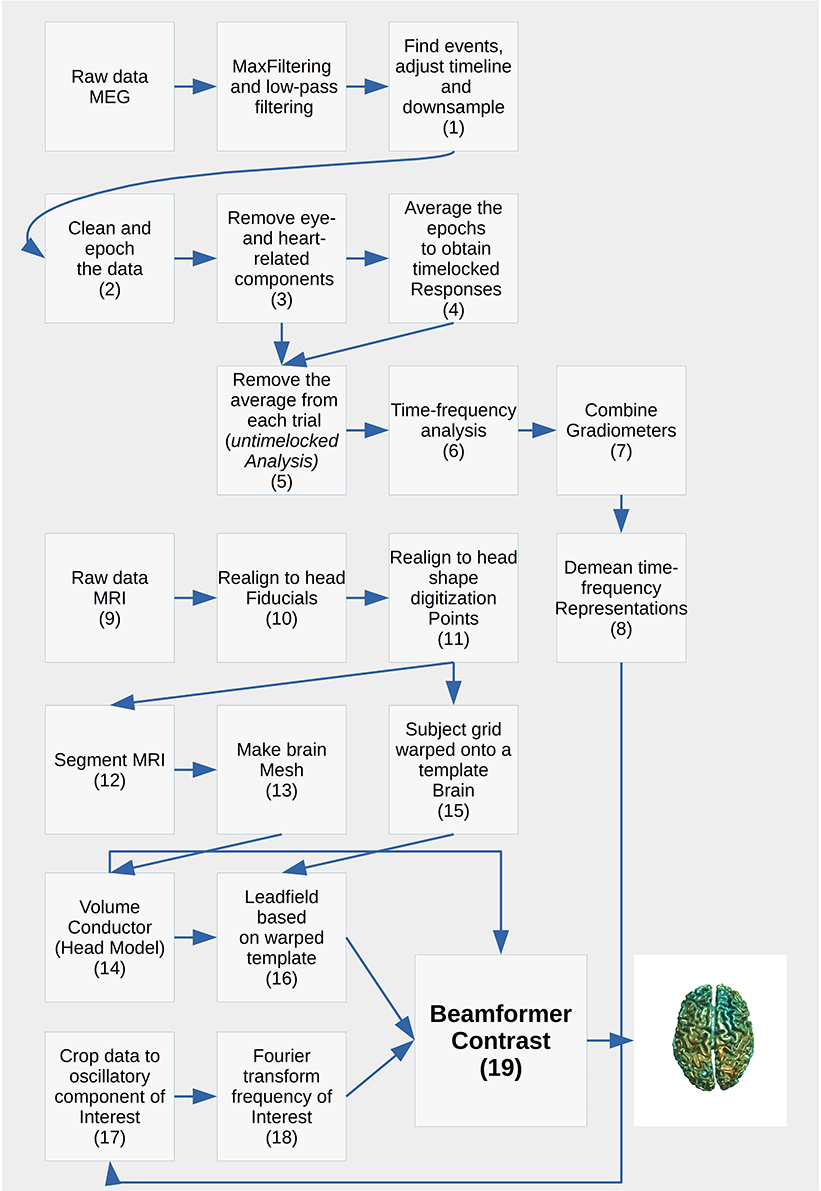
Andersen describes an analysis pipeline from raw single-subject MEG data to group-level statistics in source space. Each of the custom-written scripts has a specific purpose:
Figure copied from Andersen (2018) with permission from the author.
Still, multiple analysis steps in separate functions are required for the purpose of one script (see figure 6 for the full analysis pipeline), creating a complex hierarchy of scripts and functions:
Figure copied from Andersen (2018) with permission from the author.
To keep the computational time and storage requirements low, we applied the full analysis pipeline to two subjects only. Both the original source code from Andersen and the standardized scripts generated by reproducescript are available on GitHub. Using the documentation in Andersen (2018), we wrote the master script run_all.m , which calls all relevant functions in Andersen’s analysis pipeline in the correct order. Here we added the reproducescript option to ft_default in the same way as in the group analysis example, i.e., by initialising reproducescript separately on each subject, and then once for the group analysis (see below).
run_all.m with reproducescript enabled
This is the original code for run_all.m with the only difference that reproducescript is enabled. It is also available on GitHub.
clear
close all
global ft_default
ft_default = [];
ft_default.checksize = inf;
%% Single subject analysis
datainfo;
for do_subject = 1:numel(all_subjects)
reproduce_dir = [home_dir, sprintf('reproduce%02d/', do_subject)];
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% enable reproducescript
ft_default.reproducescript = reproduce_dir;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% Create all relevant directories where all data and all figures will be saved
create_MEG_BIDS_data_structure
% Go from raw MEG data to a time-frequency representation
sensor_space_analysis
% Go from raw MRI data to a volume conductor and a forward model
mr_preprocessing
% Extract fourier transforms and do beamformer source reconstructions
source_space_analysis
% Plot all steps in the sensor space analysis
plot_sensor_space
% Plot all steps in the MR processing
plot_processed_mr
% Plot all steps in the source space analysis
plot_source_space
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% disable reproducescript
ft_default.reproducescript = [];
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
end
%% Group analysis
datainfo;
reproduce_dir = [home_dir, 'reproduce_group'];
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% enable reproducescript
ft_default.reproducescript = reproduce_dir;
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% Do grand averages across subjects for both sensor and source spaces
grand_averages
% Do statistics on time-frequency representations and beamformer source reconstructions
statistics
% Plot grand averages in both the sensor and source spaces, with and without statistical masking
plot_grand_averages
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
% disable reproducescript
ft_default.reproducescript = [];
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%
Reproduced analysis
Please head over to GitHub to see what the reproduced script.m look like for the two subjects and for the group level.
Conclusion
In three examples we have shown how the reproducescript functionality can be applied to any analysis pipeline that is based on the FieldTrip ecosystem. This functionality can be applied without much effort on the researcher’s side, and it generates all code, original and intermediate data, as well as final results in a format in which it’s readily shareable and reproducible.
Note that there are other strategies for improving shareability and reproducibility, and we don’t claim that reproducescript is the best way in every scenario. Rather, it is one of many tools that can aid the researcher to improve the community’s standard in methodological transparency and robustness of results. For other strategies, we refer the reader to the pre-print in which we first described reproducescript.