Making your analysis pipeline reproducible using reproducescript
This example script will introduce you to functionality in the FieldTrip toolbox designed to aid in making your analysis pipeline - including code, data and results - easily reproducible and shareable. It is based on the manuscript Reducing the efforts to create reproducible analysis code with FieldTrip.
Description
Reproduciblity in neuroimaging
In recent years, unsound scientific practices have led to a replication crisis, which lead to an increased demand for greater methodological transparency and robustness in results in the scientific community. As part of this, researchers are more and more encouraged, or even obligated, by funders and journals to publish their results and analysis pipelines along with the published journal article.
The analysis of M/EEG data, and neuroimaging data in general, typically requires many calculations in which the raw data is transformed step by step. In FieldTrip and most other analysis software, these steps are written down in code in an analysis script. The more complex the analysis pipeline is, the more steps are required and the longer the analysis script becomes. Because of this, a full analysis pipeline is usually not written in a single script, but in a set of scripts and functions that are interdependent and have to be called in a certain order.
Many researchers are not formally trained as software engineers or computer scientists. Thus, the quality, readability, and generalizability of analysis scripts is highly dependent on the individual researcher’s coding style and expertise. Unfortunately, the variability in the quality of analysis scripts might compromise the reproducibility of results. Re-running the analysis pipeline might lead to different results, or the pipeline doesn’t run on someone else’s computer.
Reproducescript
In order to encourage code and data sharing, and to ensure that the shared material can reliably reproduce results, we added a functionality into FieldTrip, called reproducescript. In short: the researcher adds one additional flag to the configuration options in each FieldTrip function in the pipeline, which results in the analysis pipeline and data dependencies to be exported to a standardized representation that resembles the format of the FieldTrip tutorials. The generated scripts and corresponding data have minimal to no ambiguity. This code and data will then immediately be ready for sharing (though we encourage to add comments to the analysis scripts in order to explain what is happening).
To explain the new reproducescript functionality, we will demonstrate its use with a simple example pipeline for a single-subject analysis that comprises only a few analysis steps. Second, we demonstrate its application in a complete pipeline with preprocessing for multiple subjects, followed by a group analysis. The original idiosyncratic scripts that we selected for these first two examples are relatively clean and transparent, which means they are easily reproducible even without the new reproducescript functionality. As a final, third, example, we will apply reproducescript to an already published analysis pipeline that contains more complexity, and thus benefits more from the reproducescript functionality. The analysis code and data used in these examples are publicly available in the Donders Repository.
Example 1
To show how the reproducescript functionality works, we apply it to a script from the tutorial on Segmenting and reading trial-based EEG and MEG data. Note that before calling ft_topoplotER, we changed the units from T to fT. This is usually not done, but in this instance it serves as an example for how reproducescript handles analysis steps that were performed outside the FieldTrip ecosystem (i.e., arbitrary code).
%% LISTING 1
data_dir = '../rawdata/';
results_dir = 'analysis/';
% extract epochs
cfg = [];
cfg.dataset = fullfile(data_dir, 'Subject01.ds');
cfg.trialfun = 'ft_trialfun_general';
cfg.trialdef.eventtype = 'backpanel trigger';
cfg.trialdef.eventvalue = 3;
cfg.trialdef.prestim = 1;
cfg.trialdef.poststim = 2;
cfg = ft_definetrial(cfg);
% loading data and basic preprocessing
cfg.channel = {'MEG' 'EOG'};
cfg.continuous = 'yes'; % see https://www.fieldtriptoolbox.org/faq/continuous/
dataFIC = ft_preprocessing(cfg);
% time-lock analysis
cfg = [];
avgFIC = ft_timelockanalysis(cfg, dataFIC);
% let's make a manual change to the data that is not caputured in the provenance
avgFIC.avg = avgFIC.avg * 1e15; % convert from T to fT
% save time-locked data
save(fullfile(results_dir, 'timelock.mat'), 'avgFIC')
% plot the results
cfg = [];
cfg.xlim = [0.3 0.5];
cfg.layout = 'CTF151_helmet.mat';
ft_topoplotER(cfg, avgFIC);
% save the figure
savefig(gcf, fullfile(results_dir, 'topoplot'))
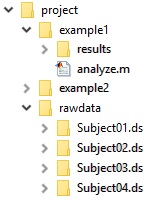
The way our files are organised can be seen in this directory tree:
The reproducescript functionality is initiated with the following code, combined with the pipeline above. The reproducescript option is enabled at the top of the script. We specify the directory to which the standard script and intermediate data are written in the reproducescript field of the global ft_default variable. ft_default is the structure in which global configuration defaults are stored; it is used throughout all FieldTrip functions and global options are at the start of the function merged with the user-supplied options in the cfg structure specific to the function. Note that we are additionally specifying ft_default.checksize = inf, which instructs FieldTrip to never remove (large) fields from any cfg-structure, thus ensuring perfect reproducibility. We recommend enabling this additional option whenever reproducescript is used.
Using reproducescript can lead to a lot of data being written to disk. Be mindful of where you save it!
clear
close all
global ft_default
ft_default = [];
ft_default.checksize = inf;
% enable reproducescript
ft_default.reproducescript = 'reproduce/';
% the original source code from listing 1 goes here.
% disable reproducescript
ft_default.reproducescript = [];
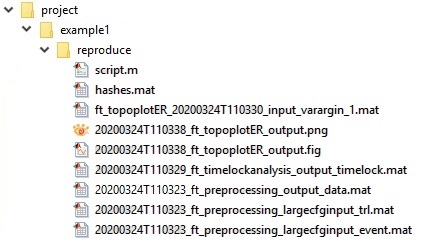
When applied to the original source code it generates a reproduce folder containing input and output data files with unique file identifiers, a MATLAB script, and a hashes.mat file:
The reproducescript functionality traces the steps to each FieldTrip function call, and recreates human-readable read-eval-print loops (REPL) code from scratch in the script.m file. For this example, script.m looks like the code below.
The reproducescript functionality copies the input data to a function and the output data it generates, and gives them a unique identifier/filename. Pointers to these identifiers end up in the standardized script as cfg.inputfile and cfg.outputfile. Note that this means that no input or output data structures as they normally appear in the MATLAB workspace appear in the standardized script; these are all handled using data on disk corresponding with cfg.inputfile and cfg.outputfile. Similarly, if the function’s output is a figure (e.g., in ft_topoplotER) the figure is also directly saved to disk in .png (bitmap) and .fig (MATLAB figure) formats.
%%
cfg = [];
cfg.dataset = '../rawdata/Subject01.ds';
cfg.trialfun = 'ft_trialfun_general';
cfg.trialdef.eventtype = 'backpanel trigger';
cfg.trialdef.eventvalue = 3;
cfg.trialdef.prestim = 1;
cfg.trialdef.poststim = 2;
cfg.tracktimeinfo = 'yes';
cfg.trackmeminfo = 'yes';
cfg = ft_definetrial(cfg);
%%
cfg = [];
cfg.dataset = '../rawdata/Subject01.ds';
cfg.trialfun = 'ft_trialfun_general';
cfg.trialdef.eventtype = 'backpanel trigger';
cfg.trialdef.eventvalue = 3;
cfg.trialdef.prestim = 1;
cfg.trialdef.poststim = 2;
cfg.tracktimeinfo = 'yes';
cfg.trackmeminfo = 'yes';
cfg.datafile = '../rawdata/Subject01.ds/Subject01.meg4';
cfg.headerfile = '../rawdata/Subject01.ds/Subject01.res4';
cfg.dataformat = 'ctf_meg4';
cfg.headerformat = 'ctf_res4';
cfg.representation = 'numeric';
cfg.trl = 'reproduce/20210112T113326_ft_preprocessing_largecfginput_trl.mat';
cfg.outputfile = { 'reproduce/20210112T113326_ft_preprocessing_output_data.mat' };
cfg.channel = {'MEG', 'EOG'};
cfg.continuous = 'yes';
ft_preprocessing(cfg);
%%
cfg = [];
cfg.tracktimeinfo = 'yes';
cfg.trackmeminfo = 'yes';
cfg.inputfile = { 'reproduce/20210112T113326_ft_preprocessing_output_data.mat' };
cfg.outputfile = { 'reproduce/20210112T113332_ft_timelockanalysis_output_timelock.mat' };
ft_timelockanalysis(cfg);
%%
% a new input variable is entering the pipeline here: 20210112T113333_ft_topoplotER_input_varargin_1.mat
cfg = [];
cfg.xlim = [0.3 0.5];
cfg.layout = 'CTF151_helmet.mat';
cfg.tracktimeinfo = 'yes';
cfg.trackmeminfo = 'yes';
cfg.inputfile = {
'reproduce/20210112T113333_ft_topoplotER_input_varargin_1.mat' };
cfg.outputfile = 'reproduce/20210112T113338_ft_topoplotER_output';
figure;
ft_topoplotER(cfg);
Note that the fields from the cfg input to ft_definetrial are repeated as input to ft_preprocessing because the configuration in the original script was not emptied. There are also additional fields created by ft_definetrial. If these fields exceed a certain printed size, which would make them unwieldy to include inline in a script (e.g., cfg.trl, which normally consists of a [Ntrials x 3] matrix specifying the relevant sections of the data on disk), these too are saved on disk instead of being printed in the standardized script. One last thing that should stand out is the comment “a new input variable is entering the pipeline here …”. This points to the mat-file subsequently specified in cfg.inputfile to ft_topoplotER. The data structure in this file was not originally created by a FieldTrip function but comes from another source: in this case it consists of the data in which originates from the T to fT unit conversion step (see the original code at the top of the page). Thus, this comment puts an emphasis on the fact that a data structure with unknown provenance enters the pipeline. See the section below on Note on using functions outside of the FieldTrip ecosystem for how to work with reproducescript and non-FieldTrip code.
Finally, the reproduce folder contains a file named hashes.mat. This is a file containing MD5 hashes for bookkeeping all input and output files. It allows reproducescript to match the output files of any one step to the input files of any subsequent step. For example, the output from ft_preprocessing is used as input to ft_timelockanalysis, which means that the data structure only needs to be stored once and xxx_ft_timelockanalysis_input_timelock.mat does not have to be additionally saved to disk. If the output data from one function and the input data to the next function are slightly different, they are both saved under different file names. This happens when the researcher modified the data using custom code (as in the example when converting channel units). The hashes.mat file furthermore allows any researcher to check the integrity of all the intermediate and final result files of the pipeline.
Using functions outside the FieldTrip ecosystem
All analysis steps that do not use FieldTrip functions will create such comments and save the data structure. Importantly, the pipeline thus remains reproducible without relying on external code. However, this does mean that it will be important to annotate script.m after it’s created and note where the data with unknown provenance comes from. Even if the pipeline exclusively uses FieldTrip functions, some FieldTrip functions evaluate custom-written code. For example, a user can specify custom code to select trials in ft_definetrial (i.e., cfg.trialfun). If this code were not shared, this particular analysis step could not be re-executed, but since intermediate results are stored as well (in the example of cfg.trialfun, cfg.trl is stored), it is always possible to skip a particular step and continue with the rest of the pipeline.
If a researcher wishes that every analysis step can be re-executed, including non-FieldTrip code, a user can “FieldTrip-ify” their non-FieldTrip functions by writing a wrapper around it (see ft_examplefunction) and Implementing a new high-level function, such that there are no unknowns in the data provenance. Under the hood, this wrapper function uses low level FieldTrip bookkeeping functions (see Toolbox architecture and organization of the source code for more information).
Conclusion
FieldTrip provides researchers with a tool to easily share complete analysis pipelines that use the FieldTrip toolbox. This is especially aimed at researchers with limited coding experience, that nevertheless want to share their analysis code and/or data with the confidence that their code is reproducible.
Here we applied reproducescript to the simplest analysis pipeline, which is example 1 in Reducing the efforts to create reproducible analysis code with FieldTrip. For more complicated analysis pipelines, have a look at example 2 and 3 in the links in the Suggested further reading.
Note that there are other strategies for improving shareability and reproducibility, and we don’t assume that reproducescript is the best way in every scenario. Rather, it is one of many tools that can aid the researcher to improve the community’s standard in methodological transparency and robustness of results. For other strategies, we refer the reader to the pre-print in which we introduce the reproducescript functionality.