How can I anonymize or deidentify an anatomical MRI?
This is something that in general you will want to do after the coregistration of the anatomical MRI with the MEG data (using ft_volumerealign), as the coregistration often relies on facial landmarks.
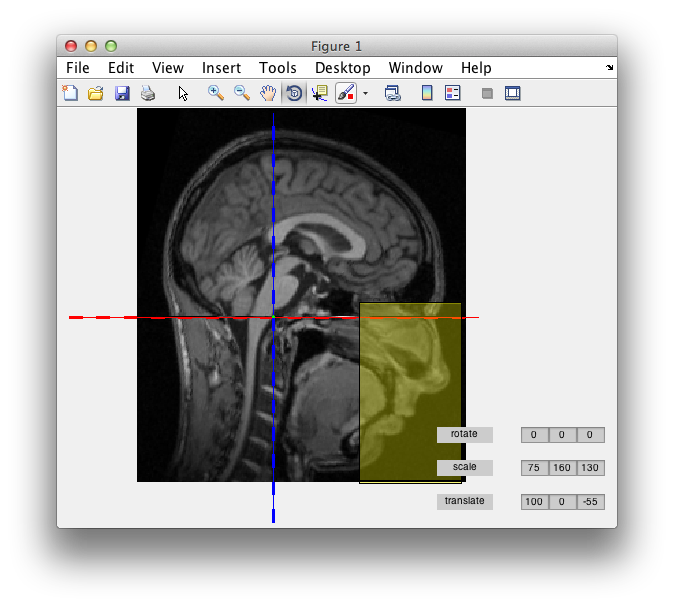
You can deface an anatomical MRI using the FieldTrip ft_defacevolume function. The default is to show a graphical user interface that allows you to scale, rotate and translate a box, such that it overlaps with the facial details that you want to be removed. Alternatively, you can use the cfg.method=’spm’ option to use an automated defacing procedure.
mri = ft_read_mri('oostenveld_r.mri');
cfg = [];
mri_anon = ft_defacevolume(cfg, mri);
You can use the standard MATLAB figure rotate button to look at the MRI from different angles.
Once you are happy with the size and position of the box, you close the figure and the function returns the defaced anatomical MRI.
You can call ft_defacevolume multiple times to sequentially mask out the identifying features. You might also want to remove the ears in two separate calls - one for each ear - as the ears are sometimes considered to be potentially identifying.
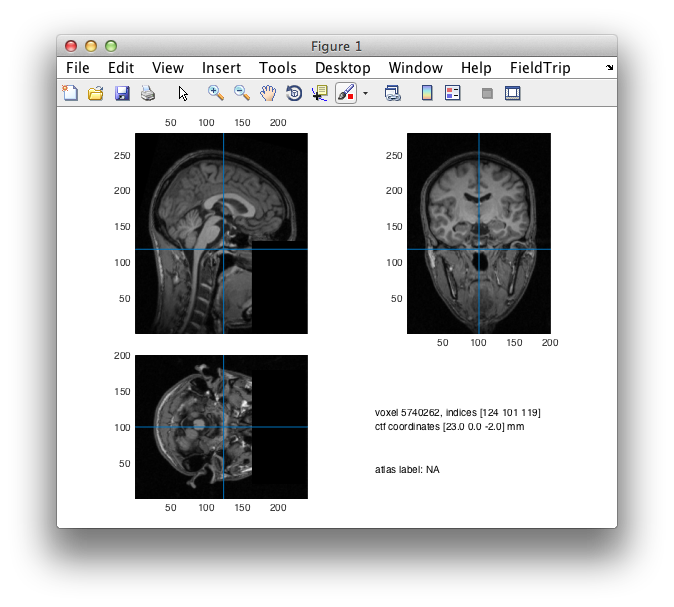
It is good practice to review the result of the defacing procedure using
cfg = [];
ft_sourceplot(cfg, mri_anon);
Subsequently you can save it to a MATLAB file or to a NIFTI file using
ft_write_mri('subjectXX_anon.nii', mri_anon.anatomy, 'transform', mri_anon.transform, 'dataformat', 'nifti');
You can also combine defacing with brain segmentation to ensure that you do not accidentally remove the orbitofrontal cortex. To do that, first we segment the anatomical MRI to extract the skull-stripped brain (including csf, white and gray matter) using ft_volumesegment. Then we inflate the brain compartment a little bit using the MATLAB imdilate function, for example, with a 5-voxel padding all around the brain. After defacing the MRI with ft_defacevolume function and possibly cutting off the orbitofrontal cortex, we can re-insert the brain anatomy back into the defaced MRI using the padded segmentation of the brain. The following script demonstrates this:
cfg = [];
cfg.output = {'brain'};
seg = ft_volumesegment(cfg, mri);
% copy the anatomy into the segmentation for plotting
seg.anatomy = mri.anatomy;
cfg = [];
cfg.funparameter = 'brain';
ft_sourceplot(cfg, seg);
% add some padding to the brain by inflating it
se = strel('sphere', 5);
seg.brain = imdilate(seg.brain, se);
cfg = [];
cfg.funparameter = 'brain';
ft_sourceplot(cfg, seg);
% we do the MRI defacing here
cfg = [];
cfg.method = 'box';
cfg.rotate = [-15 0 0];
cfg.scale = [150 75 100];
cfg.translate = [0 100 -30];
defaced_mri = ft_defacevolume(cfg, mri);
% put the brain anatomy back using the padded brain segmentation
defaced_mri.anatomy(seg.brain) = mri.anatomy(seg.brain);
cfg = [];
ft_sourceplot(cfg, defaced_mri);
See also this frequently asked question on how to anonymize a CTF MEG dataset.
If you share your MATLAB files with others, note that there might also be identifying information in the provenance information in the “cfg” field that is included in the FieldTrip data structure.
In principle FieldTrip keeps full track of all analyses that you do. The consequence might be that the original file name (identifying the subject) is included in the provenance information. You can use the ft_anonymizedata function to scrub the provenance from unwanted information.
Better is not to use the subject’s name, date of birth or other identifying information as the filename if you acquire the data. If you - or the person from whom you received the data - nevertheless did use identifying information in the file name: the earlier you rename it, the better!