Getting started with EEGLAB
Background
EEGLAB is an interactive MATLAB toolbox for processing continuous and event-related EEG, MEG and other electrophysiological data using independent component analysis (ICA), time/frequency analysis, and other methods including artifact rejection. EEGLAB incorporates and extends the ICA/EEG toolbox of Makeig, and it provides the user with a graphical interface. The homepage of EEGLAB is located at http://www.sccn.ucsd.edu/eeglab/.
How does FieldTrip use EEGLAB?
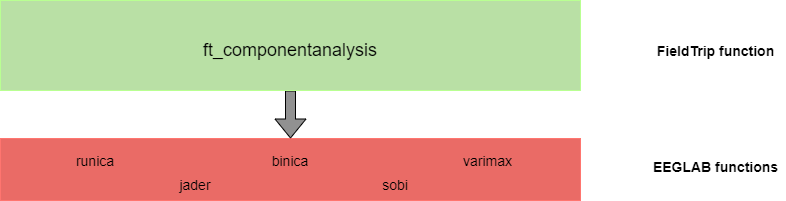
FieldTrip integrates EEGLAB functionalities to deal with component analysis. The function ft_componentanalysis directly calls EEGLAB functions to perform the desired analysis as represented in the following figure:
How does EEGLAB use FieldTrip?
EEGLAB integrates FieldTrip functions through the FILE-IO and the DIPFIT plug-ins.
The FILE-IO plug-in allows reading raw data from disk into EEGLAB, from any of the file formats that is supported by FieldTrip. Most EEG file formats are directly supported in EEGLAB, but for some MEG and iEEG file formats this plugin is needed.
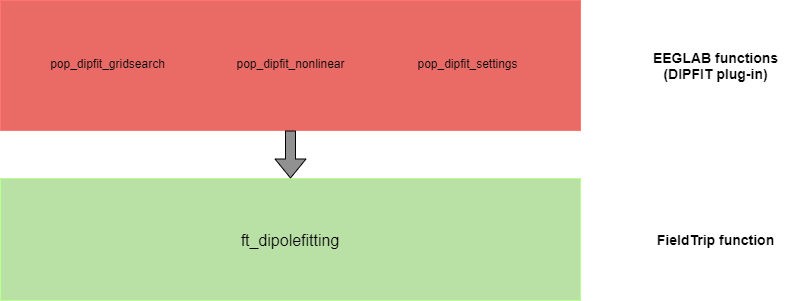
The DIPFIT plugin allows the localization of the sources that have been separated using ICA. The main function that is used by DIPFIT is ft_dipolefitting. It performs a grid search and non-linear fit with one or multiple dipoles and tries to find the location where the dipole model is best able to explain the ICA component topography, using spherical models or realistic BEM or FEM volume conduction models.
The following figure illustrates some FieldTrip functions used in DIPFIT:
Note that FieldTrip should be added to the MATLAB path in order to use the DIPFIT plug-in.
Complementary use of both toolboxes
Processing data from EEGLAB through FieldTrip functions and vice-versa is made easy by the functions eeglab2fieldtrip and fieldtrip2eeglab. Here is an example code showing how to use those functions:
-
For users working on EEGLAB, first save data on disk: File → Save current dataset (must be a
.setfile) -
(EEGLAB -> FieldTrip) Two methods can be used to import the
.setfile as a FieldTrip structure: 1. (recommended) Using ft_preprocessing:cfg = []; cfg.dataset = 'filename.set'; ft_data1 = ft_preprocessing(cfg);
- Load the dataset through pop_loadset and transform it to FieldTrip structure using eeglab2fieldtrip function:
EEG = pop_loadset(filename); ft_data2 = eeglab2fieldtrip(EEG, 'raw');
- (FieldTrip -> EEGLAB) For data that has been processed in FieldTrip, it can be converted to EEGLAB format using fieldtrip2eeglab function. Note that the data (second argument) have to be passed as an array (not a cell):
eeglab_data = fieldtrip2eeglab(ft_data.hdr,cat(3,ft_data.trial{:}));
pop_saveset(eeglab_data, 'filename', 'myfilename.set')
Notes:
-
To import frequency or timelock data to FieldTrip, ft_freqanalysis or ft_timelockanalysis can be used instead of ft_preprocessing, respectively.
-
The EEGLAB
.setand.fdtformats are directly supported by the low-level ft_read_header, ft_read_data and ft_read_event functions as well as the high-level function ft_definetrial.
Design philosophies
Both EEGLAB and FieldTrip work with data structures in MATLAB memory. The design philosophy in EEGLAB is to gather all data from one subject in a single “EEG” structure, and all data from a group of subjects in a “STUDY” structure. This is different from the design philosophy of FieldTrip, which does not gather all results in a single structure, but keeps the results from different analyses in different structures. The following example shows this philosophycal difference:
Together with the EEGLAB developers we maintain two functions for converting the data back and forth: fieldtrip2eeglab and eeglab2fieldtrip.
Releasing FieldTrip code to EEGLAB users
EEGLAB developers have created a simple tutorial so FieldTrip users can release their FieldTrip code as EEGLAB plugins. This will allow a more extensive diffusion of your methods. A template plugin also contain methods to convert data formats between the two toolboxes.