Template models for source reconstruction
Source models are a necessary ingredient for estimating the cortical activity from EEG or MEG data. Sources are typically modeled as equivalent current dipoles (ECDs), i.e. point sources with a location, orientation and strength.
You can find the template models for source reconstruction included in FieldTrip here.
Grid search in dipole fitting
When you do source reconstruction with dipole fit methods (as implemented in ft_dipolefitting), you usually assume a source model that consists of a single or a small number of equivalent current dipoles and you fit the source location, orientation and strength to the data. In this case you might start with an exhaustive grid search on a pre-defined grid, which in general is then followed by a non-linear optimization of the location of the dipoles. The initial grid on which the search is performed is not that important and is usually constructed on the fly as 3d grid with a reasonable resolution. The resolution should not be too high, otherwise the grid search will take too long, but should also not be too low, otherwise the non-lienar search will have to start from a grid location that is still far of from the minimum. Both a too low and a too high grid resolution will cause the dipole fitting procedure to take more time.
Scanning with a beamformer
When doing source reconstruction with beamformers, people typically scan the brain volume where dipoles are defined on a regular 3D grid, with a regular spacing between the dipole locations. These grids are usually optimized to the individual anatomy of the participant. To facilitate group analysis, however, a clever strategy is to use a template grid (based on a template anatomical volume) that will be (linearly or non-linearly) warped to the individual participant’s anatomy. Although this may lead to irregular spacing between the dipole locations (both across and potentially within participants), the dipole locations are directly comparable across participants, because they coincide in standard space. The FieldTrip template directory provides a set of sourcemodels defined on regular 3D-grids that are constructed from the MNI-template anatomy from SPM. These template sourcemodels can subsequently be used to be inverse normalized to the individual participant’s anatomy. See this example for more information.
Template source models with the varying dipole spacing (4, 5, 6, 7.5, 8 and 10 mm) on a regular 3-D grid are available in fieldtrip/template/sourcemodel director
- standard_sourcemodel3d10mm.mat
- standard_sourcemodel3d4mm.mat
- standard_sourcemodel3d5mm.mat
- standard_sourcemodel3d6mm.mat
- standard_sourcemodel3d7point5mm.mat
- standard_sourcemodel3d8mm.mat
To load and visualize the 3D regular grids, you can do for example
load standard_sourcemodel3d5mm.mat
figure
plot3(sourcemodel.pos(:,1), sourcemodel.pos(:,2), sourcemodel.pos(:,3), '.')
axis equal
axis vis3d
grid on
or you can use
figure
ft_plot_mesh(sourcemodel)
You will notice that the 3D regular grids are not that interesting to look at; since there are no connecting elements (triangles), ft_plot_mesh will only show the vertices.
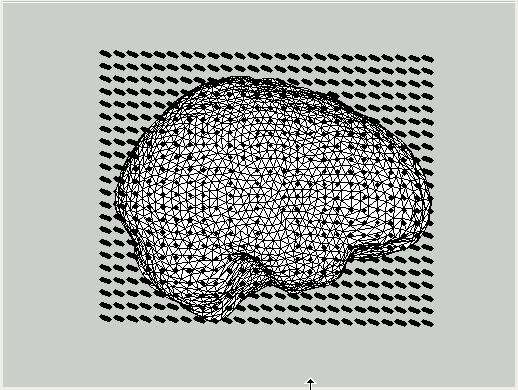
Distributed source models with MNE
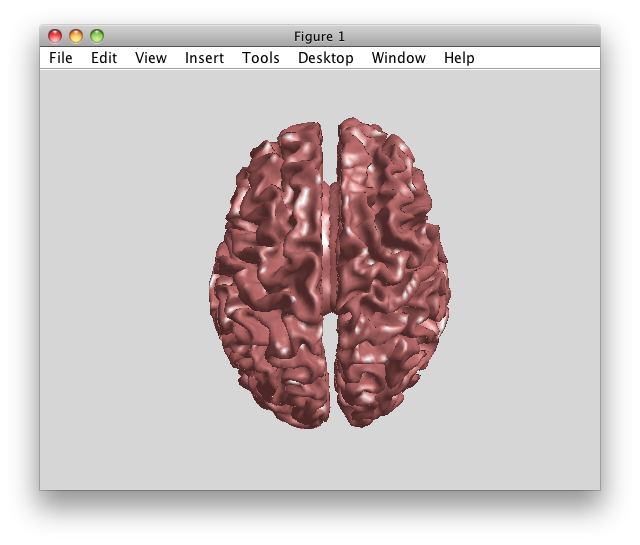
When doing source reconstruction using minimum norm estimation (MNE, also known as linear estimation) techniques, the assumption is that the sources in the brain are distributed and that only the strength at all possible cortical locations is to be estimated. Since the strength of all dipoles in the cortical mesh is estimated simultaneously, sources should only be placed in regions where generators might be present. MNE therefore usually assumes a source model that consists of grey matter only, which can be modeled as a highly folded cortical sheet.
A canonical cortical sheet is available in fieldtrip/template/sourcemodel with different numbers of vertices (20484, 8192 and 5124 vertices). These files were taken from the SPM8 release version; they refer to the canonical T1 anatomical MRI and are expressed in MNI coordinates.
- cortex_20484.surf.gii
- cortex_8196.surf.gii
- cortex_5124.surf.gii
You can load and visualize the cortical sheets with
sourcemodel = ft_read_headshape('cortex_20484.surf.gii')
ft_plot_mesh(sourcemodel, 'facecolor', 'brain', 'edgecolor', 'none')
camlight
lighting gouraud
UPDATE: cortical meshes from SPM8 added on 17 May, 2013
The following documentation is a verbatim copy of spm_mesh.man in SPM8 and describes the details of
- cortex_20484.surf.gii
- cortex_5124.surf.gii
- cortex_8196.surf.gii
The cortical mesh surfaces here were created using FreeSurfer version 4.0.1 (Dale et al. 1999, Fischl et al. 2001, Fischl et al. 1999) from an average of 27 T1 scans of the same subject (see spm_templates.man and Tzourio-Mazoyer et al. 2002 for more details). The surfaces were inflated to a sphere and down-sampled using an octahedron (8,196 vertices) or an icosahedron (5,124 and 20,464 vertices) equally subdivided to achieve the highly tessellated surfaces provided.
The boundary element model surfaces were created using the watershed algorithm (Segonne et al. 2004). These surfaces were created utilizing an icosahedron equally subdivided to create a highly tessellated surface. Some manual editing was applied to the T1 images in order to optimize the performance of the watershed algorithm.
The meshes were subsequently warped using the deformation field created by the new segmentaton algorithm, so that the reference space was defined by spm8/toolbox/Seg/TPM.nii, to improve the accuracy of the alignment.
They are saved in the GIfTI file format, with GZipBase64Binary encoding.
References
- Dale, A.M., Fischl, B., Sereno, M.I., 1999. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 9, 179-194.
- Fischl, B., Liu, A., Dale, A.M., 2001. Automated manifold surgery constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging 20, 70-80.
- Fischl, B., Sereno, M.I., Dale, A.M., 1999. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage 9, 195-207.
- Segonne, F., Dale, A.M., Busa, E., Glessner, M., Salat, D., Hahn, H.K., Fischl, B., 2004. A hybrid approach to the skull stripping problem in MRI. Neuroimage 22, 1060-1075.
- Tzourio-Mazoyer, N., Landeau, B., Papathanassiou, D., Crivello, F., Etard, O., Delcroix, N., et al, 2002. Automated anatomical labelling of activations in spm using a macroscopic anatomical parcellation of the MNI MRI single subject brain. Neuroimage 15, 273-289.
UPDATE: new 3D source models uploaded on March 7, 2013
Up until March 7, 2013 (svn revision number 7600) the sourcemodels were a bit tight at the top of the brain. This means that for most of the individual subjects (after warping the template grid from standardized space into individual space) the very top of the brain was not fully covered by dipoles. This is demonstrated in figure 1 below. The reason for this turned out to be the fact that we used the blurry T1.nii template MRI to make a surface description of the brain (determining the inside and outside dipoles). This turned out to be too tight for most individual (non-blurred) anatomicals, see figure 2. We therefore uploaded a new set of sourcemodels to be released with FieldTrip as of March 7, 2013. These new sourcemodels provide a looser fit to the template brain surface. The template brain surface was extracted using a 1 mm resolution non-blurred template MRI. An example of the updated sourcemodel, warped to an individual brain is shown in figure 3.
The new set of sourcemodels are not compatible with the old set. If you were in the middle of an analysis, that relies on the template sourcemodels in FieldTrip, you should either stick to the old version of the sourcemodels, or recompute all results using the new version of the sourcemodels. The old version of the sourcemodels will not be kept in the release version of FieldTrip, but they are recoverable from FieldTrip version predating March 7, 2013.
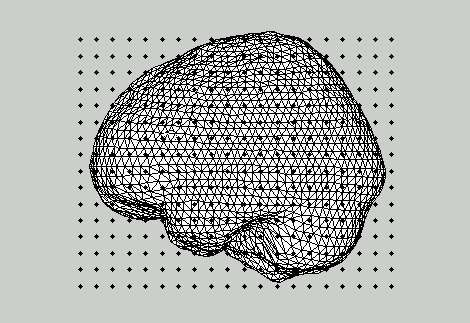
Figure 1A: Template brain surface with template dipole locations (old version sourcemodel).
Figure 1B: Warped dipole locations (linear warp only) on top of an individual brain surface.
Figure 1C: Warped dipole locations (nonlinear warp) on top of an individual brain surface.
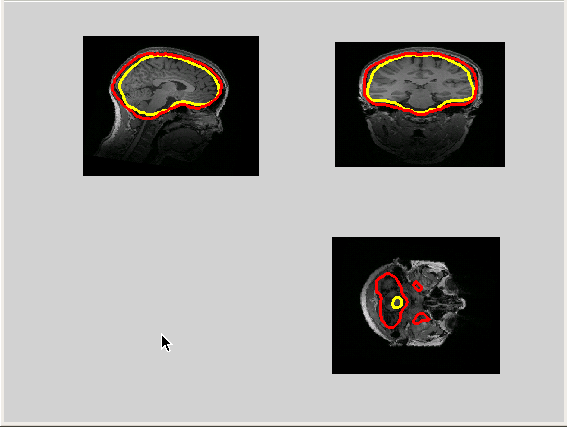
Figure 2: The brain surface extracted from the blurred template brain warped to, and projected onto an individual MRI (yellow), and the brain surface extracted from the individual MRI (red).
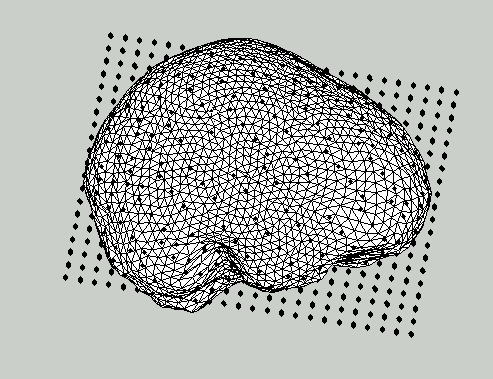
** Figure 3: Updated sourcemodel with template dipole locations (linearly warped) on top of an individual brain surface.**
UPDATE: updated 3D source models uploaded on February 19, 2016
Essentially this update does not include a functional change, it only pertains to the structure in the data-structures. Before, the source model variable contained ‘xgrid’/’ygrid’/’grid’ fields. These fields since long have been obsolete. Also, the inside/outside handling used to be in terms of index-vectors. More recently, the default functionality has been replaced by inside being a boolean vector, which is easier to handle in terms of bookkeeping. The sourcemodels now reflect these current standards.