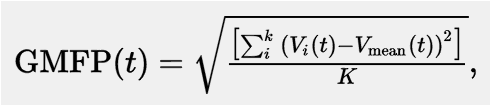Dealing with TMS-EEG datasets
Introduction
This tutorial shows how to process EEG that was recorded together with transcranial magnetic stimulation (TMS) that was applied to the primary motor cortex (M1), while subjects either contracted, or relaxed their contra-lateral hand. The application of TMS pulses during the EEG acquisition poses some specific challenges that will be addressed.
Dealing with the TMS-artifacts is best done as a preprocessing step prior to starting the scientifically interesting parts of the analysis. Once the TMS artifacts have been removed, you can proceed with the EEG analysis pipeline as usual. This tutorial shows some analysis results, but does not elaborate on the EEG analysis methods in general.
The research question that we will address in this tutorial is whether pre-contraction of the hand affects the TMS-evoked potential (TEP) in the EEG. To answer this question we will look at
- TEP components
- Frequency content of spontaneous oscillations
- global mean field power
Background
A successful analysis of EEG signals requires clean data. That is non-trivial for EEG in general, but the TMS induced artifacts make it an even bigger challenge. As with all other artifacts, prevention is better than cure. In combined TMS-EEG experiments it is however unavoidable that some TMS artifacts appear in your data. The use of proper equipment (amplifiers, electrodes, etc) and fine-tuning of acquisition settings can help reduce the artifacts and facilitate the analysis. However, optimizing the acquisition is not the topic of this tutorial; please see the ‘suggested reading’ section at the end for more information on this.
The dataset used for this tutorial
The data used in this tutorial is available from our download server where you can find a single-pulse dataset (used here) and a paired-pulse dataset.
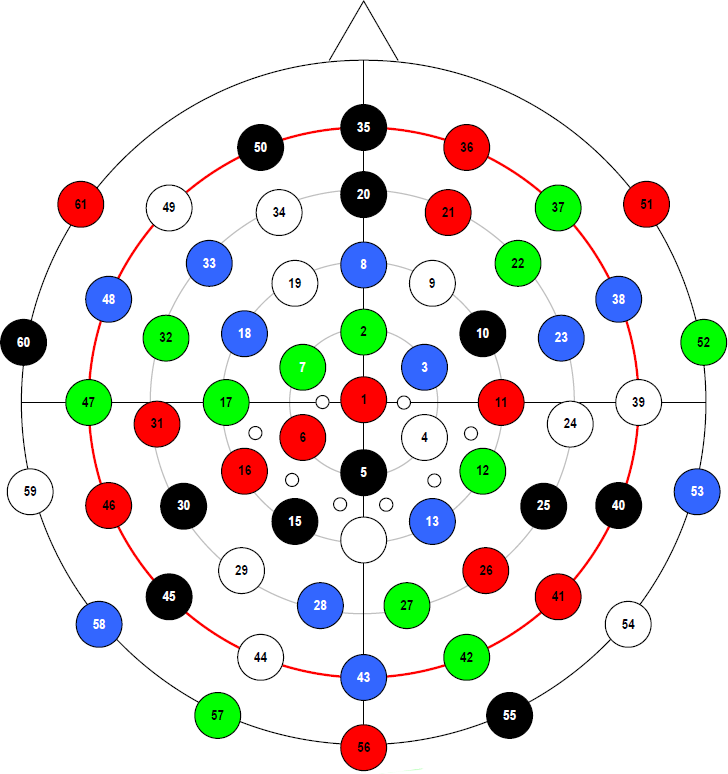
Measurement: The data were recorded using two 32-channel TMS-compatible BrainAmp DC Amplifiers (BrainProducts) connected to a 61 channel TMS-compatible EEG cap (EasyCap). Sampling was done at 5kHz with a 1kHz cut-off frequency and with 0.1 microvolt/bit resolution. Please click on the image below for an enlarged image of the equidistant 61-channel arrangement.
TMS: Single pulses were applied at 60% maximum stimulator output using a C-B60 butterfly coil connected to a MagPro X100 (Magventure) stimulator. In total 300 pulses were applied with an inter-pulse-interval of 3 seconds (20% jitter).
The coil position was determined by finding the motor hotspot. The stimulation location was kept constant using MRI-guided neuronavigation (Localite). For the purpose of this tutorial the orientation and tilt of the coil were adjusted to induce strong artifacts.
Event markers: An event was placed at the onset of each TMS-pulse indicating the condition. ‘S 1’ represents the ‘Relax’ condition and ‘S 3’ represents the ‘Contract’ condition.
Memory Issues
In TMS-EEG research we are often dealing with large datasets due to high sampling rates and lengthy recording sessions. It is therefore quite common that data structures loaded into MATLAB’s memory are multiple gigabytes in size.
The maximum size a MATLAB array can be depends on the operating system, the MATLAB version and the amount of RAM. If you are running a 32-bit version of Windows, your arrays cannot be larger than 2 gigabytes. If you are running a 32-bit version of MATLAB on a 64-bit version of Windows your arrays can still not be larger than 4 gigabytes. Only when you are running both a 64-bit version of MATLAB and Windows, can you use the full extent of your RAM and are theoretically able to store arrays as large as 8 terabytes.
We therefore advise you to run this tutorial on a 64-bit operating system, running a 64-bit version of MATLAB and with at least 8GB RAM.
Also see: Resolving “Out of Memory” Errors
TMS paradigm
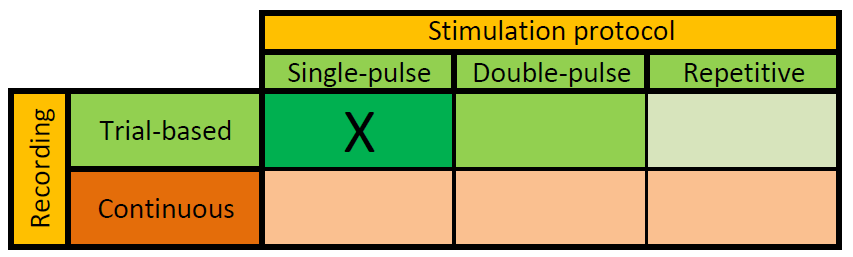
Before starting your analysis, it helps to consider your experiment relative to two dimensions: 1) the TMS protocol and 2) the experimental manipulation of the brain state (i.e. the task for the subject, or the absence thereof).
In many cases you will want to analyze your data with respect to some interesting event, e.g., a visual stimulus onset, the subjects response to a stimulus, or in this case the onset of a TMS pulse. It is, however, also possible to analyze your data in a continuous way. For example, you might be interested in resting-state EEG changes over time after participants have received theta-burst stimulation. In those cases you are not going to divide your data into experimentally defined trials but rather analyze the data in a continuous fashion.
This tutorial is written with a trial-based analysis in mind and as such may not be directly applicable to continuous data. Furthermore, this tutorial uses EEG data recorded during a single-pulse TMS stimulation protocol.
It is also important to realize that between datasets the amount and spacing of the pulses will vary. In principle we can distinguish between one pulse per trial, two-pulses per trial, or repetitive stimulation. Although this tutorial was written for single-pulse studies, most of it also applies to multiple-pulse data.
Artifacts
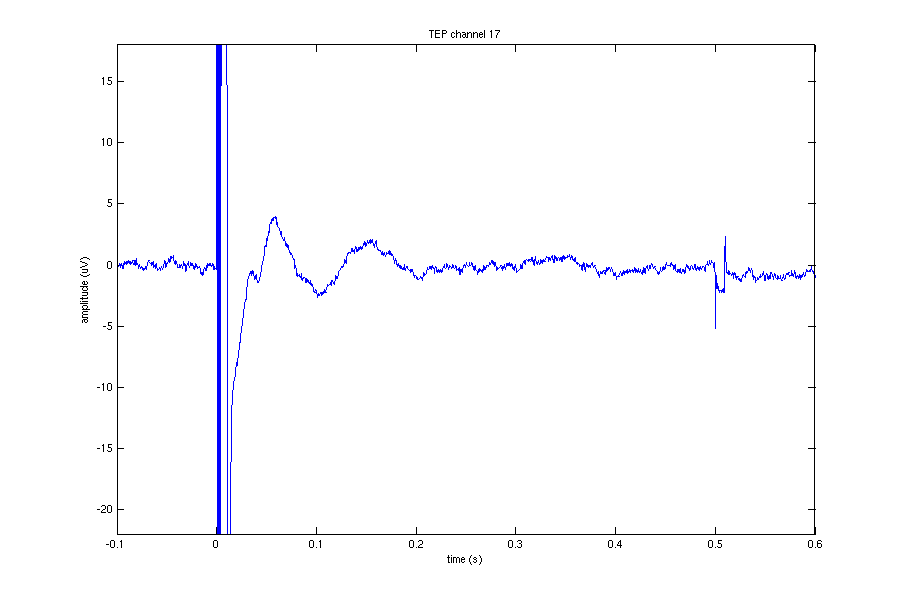
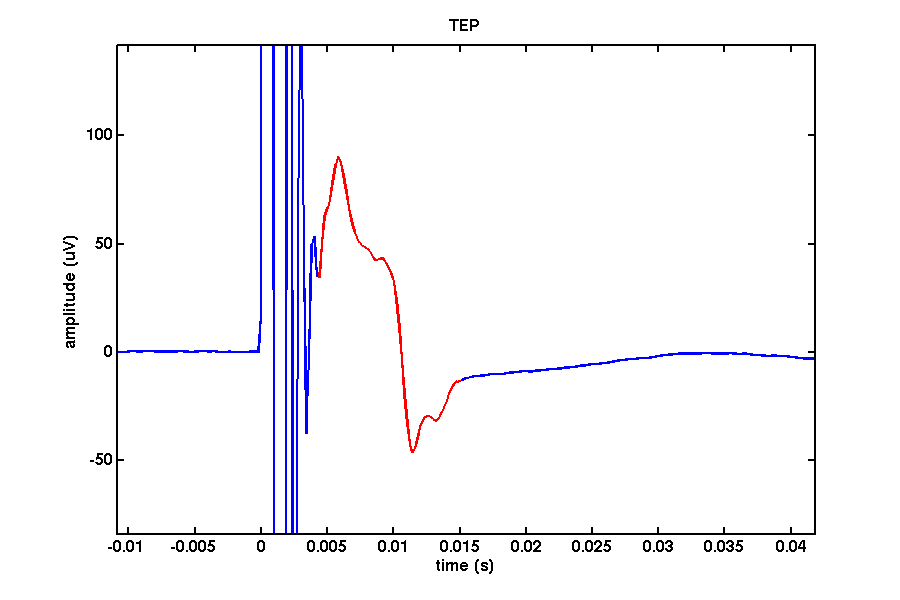
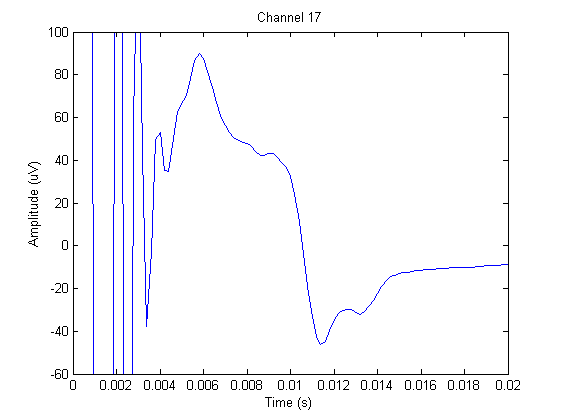
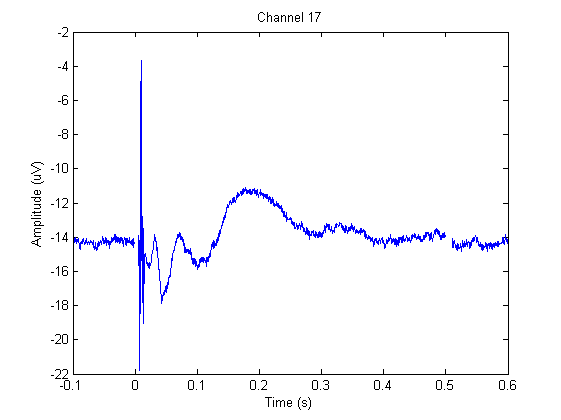
The figure below shows the EEG on channel 17 (see the layout of electrodes above) during the application of the TMS.
We will now shortly describe and display the TMS-related artifacts you can come across in your data. It is important to understand that there is not just a single type of artifact, different artifacts may occur simultaneously. Depending on the characteristics of EEG and TMS equipment and experimental design, not all types of artifacts may be observed in your data.
Pulse artifact
The data during the pulse can be considered to be lost.
Ringing/Step response artifact
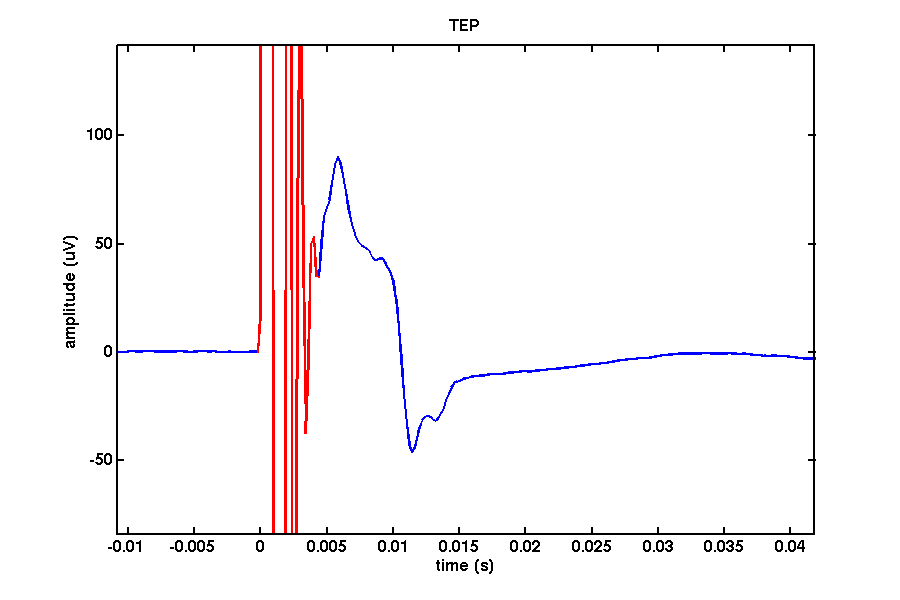
Depending on the range of your amplifier the signal may initially go out of range causing a clipping of the signal as can be seen by a flat line in your signal. Once the potential falls within range of the amplifier a prominent filter ‘ringing’ can be observed lasting up to around 7ms depending on your setup caused by a step-response due to the high gradient of the TMS-pulse. Many consider this period lost. In the figure below the signal in red reflects a combination of the pulse and the ringing/step response.
Cranial muscle artifact
The TMS pulse may cause cranial (scalp) muscle twitches. These twitches are not to be confused with responses due to stimulation of the motor cortex but are purely twitches due to stimulation of scalp muscles. Usually they last around 10ms and are orders of magnitude larger than brain signals. Using close visual inspection of the EEG electrodes underneath the TMS coil during the actual experiment, you may observe these twitches as small movements of the electrodes.
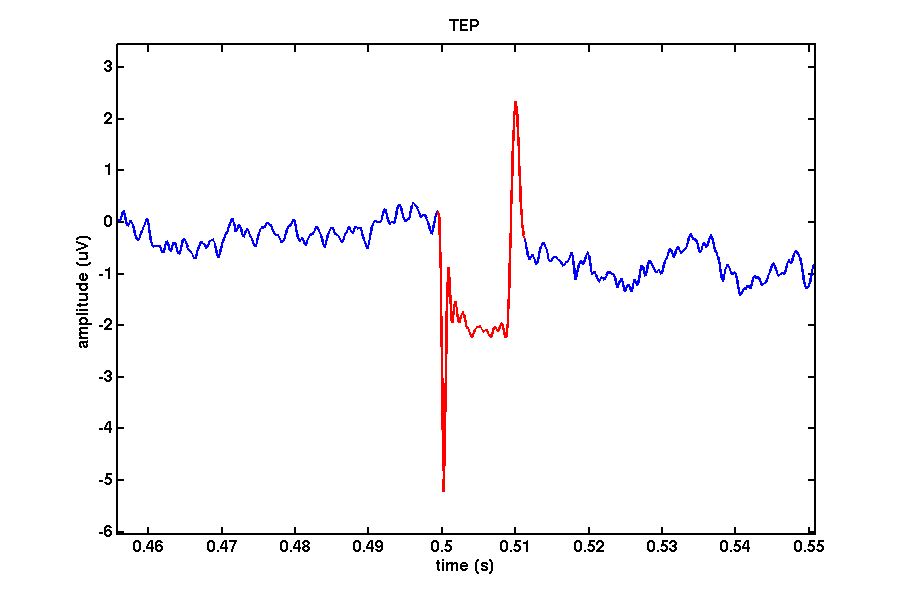
Recharging artifact
Depending on your TMS machine you may observe a spike in your data reflecting the recharging of your machine’s capacitors. Some stimulators allow you to specify the exact time the machine recharges its capacitors. In the case of this dataset we used a MagPro X100 (Magventure) stimulator with the recharge delay set to 500ms after stimulation onset.
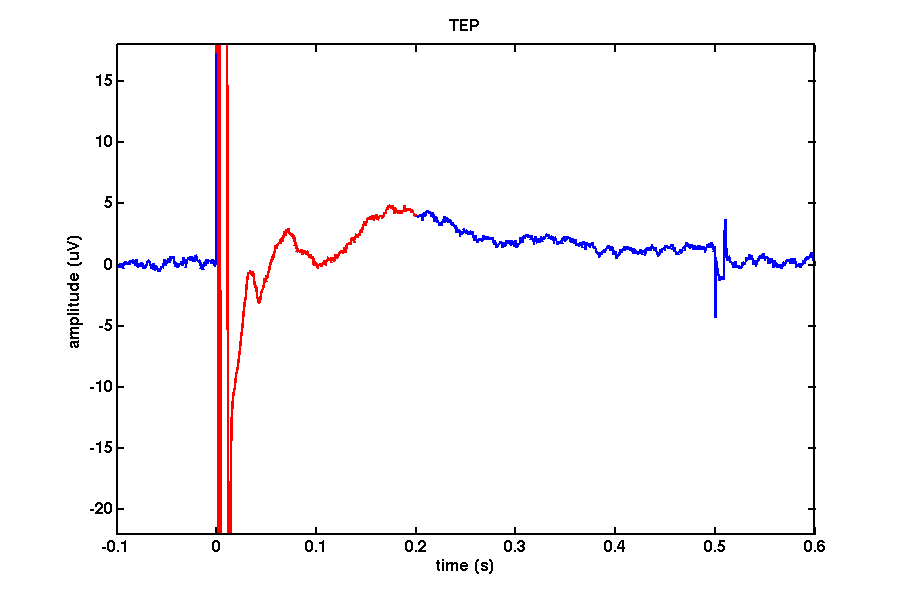
Decay artifact
It is likely that you will encounter an artifact that resembles an exponential decay in some channels. The nature of this artifact is not well understood and its presence is rather unpredictable. We believe that it arises due to an interaction between magneto-electric induction of TMS-induced currents in the electrode leads, electrode-electrolyte-skin interface polarization, movement of the electrodes and head/neck/face muscle twitches. In worst cases, this artifact can last up to 1 second, but more commonly it lasts between 50-150 ms.
Procedure
The procedure consists of preprocessing (and data cleaning), followed by the computation of Event Related Potentials (ERPs) and time-frequency responses (TFRs). The analysis in this tutorial was used for and is described in more detail in the following paper:
Herring, J. D., Thut, G., Jensen, O., & Bergmann, T. O. (2015). Attention Modulates TMS-Locked Alpha Oscillations in the Visual Cortex. The Journal of Neuroscience, 35(43), 14435-14447.
We will use the following procedure for preprocessing the TMS-EEG data:
- Create a trial-structure using ft_definetrial
- Indicate the onset of TMS-pulses with ft_artifact_tms
- Visually determine which artifacts are present using ft_preprocessing, ft_timelockanalysis, and ft_databrowser.
- Exclude ringing/step response and recharge artifacts from the trial-structure using ft_rejectartifact and ft_artifact_tms
- Read-in segments excluding previously rejected artifacts using ft_preprocessing.
- Perform Independent Component Analysis to attempt to remove exponential decay and cranial muscle artifacts using ft_componentanalysis and ft_rejectcomponent. At this stage independent components related to other artifacts (e.g., line noise, eye blinks/saccades) can be removed as well.
- Recreate the intended trial structure using ft_redefinetrial
- Interpolate gaps previously occupied by ringing/step response and recharge artifacts using ft_interpolatenan
- Apply further processing such as baseline correction, detrending, and filtering using ft_preprocessing.
After having preprocessed and cleaned the data, we will perform the following analyses:
- Calculate time-locked averages using ft_timelockanalysis
- Time-frequency analysis using ft_freqanalysis
- Calculate the global mean field power
Preprocessing
It is important that you clean your data from TMS artifacts prior to any other preprocessing steps (e.g., filtering, detrending, downsampling) other than reading the data into memory. Especially filtering can produce long-lasting additional artifacts far outlasting the duration of the artifacts in the raw data, making subsequent analysis of the data troublesome.
Visual data inspection
We start with the original dataset in BrainVision format which is available here. Please be aware that the file is rather large (472 MB) due to the EEG being sampled at 5kHz.
We are interested in what happens in response to the TMS pulse. The TMS pulses are therefore our events of interest and our trials are defined by the pulses. As stated in the background information, event markers are present at the onset of each pulse. We will first have a look at our trials using ft_databrowser, a convenient tool to browse data directly from disk or in memory (also see this frequently asked question).
The complete dataset is rather memory demanding, hence we will only read the segments of interest (i.e. the trials) from disk using ft_preprocessing. For this purpose we first need to create a trial matrix, which specifies which parts of the data on disk are to be represented as trials. This matrix has three (or more) columns and as many rows as there are trials. The first two columns indicate the sample number in the data file on disk corresponding to the first and last sample of each trial. The third column reflects the so-called offset, i.e. which sample corresponds to time point zero in each trial. The matrix can have additional columns containing information, such as the experimental condition for each trial, the reaction time, etc. The trial matrix is created using ft_definetrial as explained in the Preprocessing - Segmenting and reading trial-based EEG and MEG data tutorial.
triggers = {'S 1', 'S 3'}; % These values correspond to the markers placed in this dataset
cfg = [];
cfg.dataset = 'jimher_toolkit_demo_dataset_.eeg';
cfg.continuous = 'yes';
cfg.trialdef.prestim = .5; % prior to event onset
cfg.trialdef.poststim = 1.5; % after event onset
cfg.trialdef.eventtype = 'Stimulus'; % see above
cfg.trialdef.eventvalue = triggers ;
cfg = ft_definetrial(cfg); % make the trial definition matrix
The output cfg variable contains the trial structure in cfg.trl. As we will also need this trial structure later in this tutorial, we will copy it into another MATLAB variable.
trl = cfg.trl;
The cfg structure we obtained from ft_definetrial contains enough information for ft_preprocessing to read our data from disk into trials. We will, however, also specify that the data should be re-referenced. As it can take quite a while (5-10 minutes) to read-in the data, the processed data can be found here. If you have downloaded this file, you can load the data with:
load data_tms_raw
Here you have to make sure that the .mat file is located in the present working directory of MATLAB.
You can skip the following block of code if you have downloaded data_tms_raw.mat in the previous step.
To read the trials from the original data file on disk, use the following:
cfg.channel = {'all' '-5' '-mastoid L' '-mastoid R'}; % indicate the channels we would like to read and/or exclude.
cfg.reref = 'yes'; % We want to rereference our data
cfg.refchannel = {'all'}; % Here we specify our reference channels
cfg.implicitref = '5'; % Here we can specify the name of the implicit reference electrode used during the acquisition
data_tms_raw = ft_preprocessing(cfg);
The reference electrode used during the acquisition is not present in the data file, however, we know that the potential (per definition) at that electrode is zero Volt. We add this “implicit” reference channel (implicit, as it is not specified in the data file) to the analysis by specifying its name.
If you have decided to read the trials from the data file on disk, use the following code to save the processed data structure for future use.
save('data_tms_raw','data_tms_raw','-v7.3');
We will now visually inspect the data using ft_databrowser. For plotting purposes we will apply a baseline correction using the pre-stimulation period.
cfg = [];
cfg.preproc.demean = 'yes';
cfg.preproc.baselinewindow = [-0.1 -0.001];
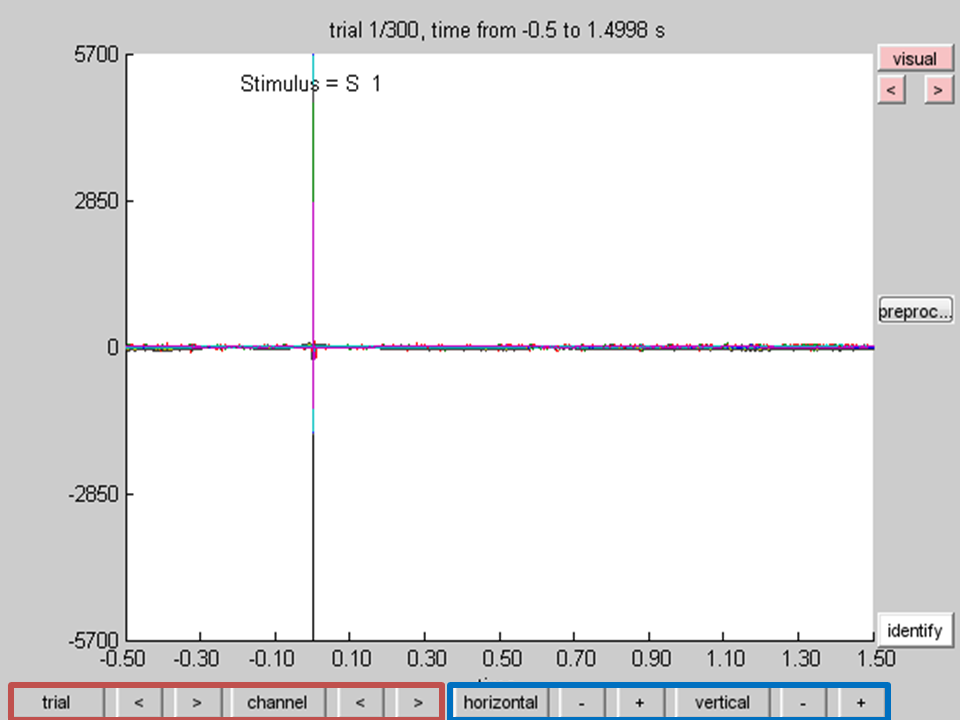
ft_databrowser(cfg, data_tms_raw);
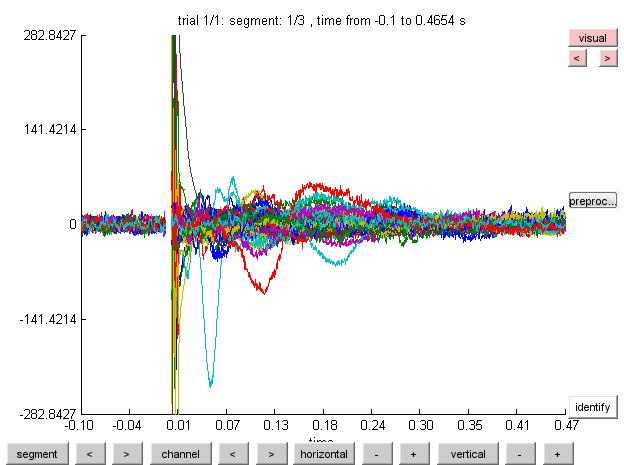
It is important to get a feeling for the quality of your recording prior to your analysis. You will have to adjust the scaling, as the amplitude of the TMS pulse is enormous compared to the actual EEG signal. Using the + and - next to the ‘vertical’ and ‘horizontal’ button (marked in blue) you can adjust the scaling on both axes. Take a moment to browse through the trials with the arrow buttons next to the trial button.
Note that if you adjust the horizontal scaling, the trial button changes to a segment button, indicating that you are browsing segments within one trial. If you want, you can make a selection of the channels you wish to plot by clicking on the ‘channel’ button (marked in red). If you want to know which channel represents which line in the plot, click on the ‘identify’ button and click on a line in the plot. The corresponding channel will be displayed above the line. Browsing through the trials you will notice a lot of noise. Furthermore, it appears that around 500 ms into the trial saccade signals appear. We can also see eye-blinks throughout the trials. At this point it is possible to mark data segments for rejection by clicking and dragging any area within the trial and clicking on the selection. You can later reject the specific segment, remove the entire trial, or replace it with nans. For now this step will be skipped as we will try to remove the physiological artifacts with Independent Component Analysis (ICA).
To inspect the TMS-related artifacts we will create a time-locked average of our data. When TMS-EEG artifacts occur, they will occur in every trial at the same time, which makes it easier to spot them in the time-locked averages.
cfg = [];
cfg.preproc.demean = 'yes';
cfg.preproc.baselinewindow = [-0.1 -0.001];
data_tms_avg = ft_timelockanalysis(cfg, data_tms_raw);
To save system memory we will clear the data_tms_raw structure.
clear data_tms_raw
Although we do not have any trials anymore, we can still use ft_databrowser in the same way as before to browse through all the channels. As we are interested in the occurrence, onset, and offset of TMS artifacts it as convenient to use MATLAB’s built-in plotting functions.
The averaged data we want to plot is represented in the data_tms_avg structure. Lets have a look at this structure:
>> data_tms_avg
data_tms_avg =
avg: [61x10000 double]
var: [61x10000 double]
time: [1x10000 double]
dof: [61x10000 double]
label: {61x1 cell}
dimord: 'chan_time'
cfg: [1x1 struct]
Time is represented in the .time field, the amplitudes are stored in .avg. When inspecting this .avg field we can see that it has a dimension of 61x10000. In this case you may have guessed that the rows represent the channels and the columns the time-points within each channel. If you are uncertain about the order of the dimensions you can always look at the .dimord field. The .dimord field tells you what the dimensions in the data field represent. Here we see that the dimord is chan_time. Therefore, our data field (.avg) is represented by channels X time. Channel labels can be found as strings located in the .label field.
In our case the electrode labels are equal to the channel numbers. Please be aware that this does not necessarily correspond to the row-numbers in the data matrices. For example, our reference electrode used in the recording is number 5 on the cap. This channel, however, is now represented in the last row (61) of the data matrix. Always check the .label field of your data structure which row of the data corresponds to which channel and electrode.
We will now plot the data for all channels in separate windows. You can use the following code to close all previous figure window
close all
for i=1:numel(data_tms_avg.label) % Loop through all channels
figure;
plot(data_tms_avg.time, data_tms_avg.avg(i,:)); % Plot this channel versus time
xlim([-0.1 0.6]); % Here we can specify the limits of what to plot on the x-axis
ylim([-23 15]); % Here we can specify the limits of what to plot on the y-axis
title(['Channel ' data_tms_avg.label{i}]);
ylabel('Amplitude (uV)')
xlabel('Time (s)');
end
Take a moment to have a look at all channels. See if you can determine the artifacts that are present.
We will have a closer look at channel 17, which is close to the site of stimulation.
channel = '17';
figure;
i = find(strcmp(channel, data_tms_avg.label));
plot(data_tms_avg.time, data_tms_avg.avg(i,:)); % Plot data
xlim([-0.1 0.6]); % Here we can specify the limits of what to plot on the x-axis
ylim([-23 15]); % Here we can specify the limits of what to plot on the y-axis
title(['Channel ' data_tms_avg.label{i}]);
ylabel('Amplitude (uV)')
xlabel('Time (s)');
If we adjust the plotting limits a bit, we can easily distinguish the ringing/step response (~0-0.0045) from the cranial muscle (~0.0045 - 0.015). You can also do this manually using the zoom button or with
xlim([-0 0.020]);
ylim([-60 100]);
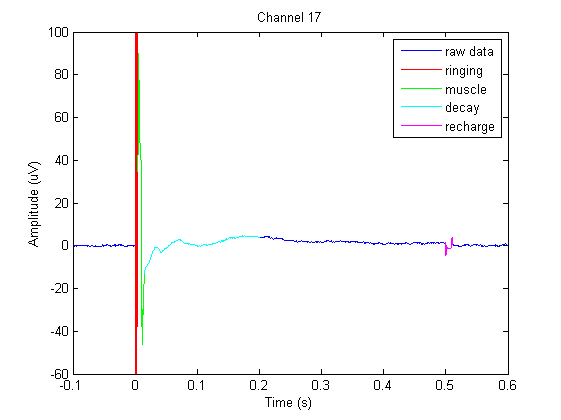
In this channel we can find ringing/step response, cranial muscle, exponential decay, and recharging artifacts. The following code highlights these artifacts in this channel.
channel = '17';
figure;
channel_idx = find(strcmp(channel, data_tms_avg.label));
plot(data_tms_avg.time, data_tms_avg.avg(channel_idx,:)); % Plot all data
xlim([-0.1 0.6]); % Here we can specify the limits of what to plot on the x-axis
ylim([-60 100]); % Here we can specify the limits of what to plot on the y-axis
title(['Channel ' data_tms_avg.label{channel_idx}]);
ylabel('Amplitude (uV)')
xlabel('Time (s)');
hold on % Plotting new data does not remove old plot
% Specify time-ranges to higlight
ringing = [-0.0002 0.0044];
muscle = [ 0.0044 0.015 ];
decay = [ 0.015 0.200 ];
recharge = [ 0.4994 0.5112];
colors = 'rgcm';
labels = {'ringing','muscle','decay','recharge'};
artifacts = [ringing; muscle; decay; recharge];
for i=1:numel(labels)
highlight_idx = [nearest(data_tms_avg.time,artifacts(i,1)) nearest(data_tms_avg.time,artifacts(i,2)) ];
plot(data_tms_avg.time(highlight_idx(1):highlight_idx(2)), data_tms_avg.avg(channel_idx,highlight_idx(1):highlight_idx(2)),colors(i));
end
legend(['raw data', labels]);
Exercise: find artifacts in other channels
Try to see if the artifacts are present in all channels and if there are differences in their extent in time.
Artifact exclusion
In this part of the tutorial we are going to exclude artifactual data segments that cannot be attenuated by other methods. We will exclude the segments that contain the ringing/step response artifact and the recharging artifact. We will adjust the trial structure of our data, containing the information which samples correspond to which trials, so that it does not include these artifacts and we will then read-in the data without these artifactual parts. Later on we will interpolate gaps which are produced by excluding these segments. The interpolation of the missing data segments is postponed until after applying independent component analysis in the next part of the tutorial.
The function ft_rejectartifact can adjust the trial structure to exclude segments containing artifacts. We first have to tell it which segments to exclude. For this purpose we will use ft_artifact_tms. This function can be used to either detect TMS pulses in your data or to specify the onset of TMS pulses by use of marker information.
% Ringing/Step Response artifact
trigger = {'S 1','S 3'}; % Markers in data that reflect TMS-pulse onset
cfg = [];
cfg.method = 'marker'; % The alternative is 'detect' to detect the onset of pulses
cfg.dataset = 'jimher_toolkit_demo_dataset_.eeg';
cfg.prestim = .001; % First time-point of range to exclude
cfg.poststim = .006; % Last time-point of range to exclude
cfg.trialdef.eventtype = 'Stimulus';
cfg.trialdef.eventvalue = trigger ;
cfg_ringing = ft_artifact_tms(cfg); % Detect TMS artifacts
In ft_artifact_tms we use cfg.prestim and cfg.poststim to indicate the time range around the TMS pulse that should be removed. There is no restriction, however, that prevents both time points from being located after the pulse (in case of a recharging artifact, for example). It is therefore also possible to mark artifacts that have on- and offsets both after onset of the TMS pulse.
In case you wish to specify an artifact onset that occurs after the TMS pulse (e.g., in case of a recharging artifact), cfg.prestim must be negative (e.g., -0.500 for 500 ms after pulse onset) as cfg.prestim refers to time before stimulus onset.
% Here we use a negative value because the recharging artifact starts AFTER TMS-pulse onset
cfg.prestim = -.499;
cfg.poststim = .511;
cfg_recharge = ft_artifact_tms(cfg); % Detect TMS artifacts
We now have a configuration structure for both the ringing/step response artifact as well as the recharging artifact. The ft_artifact_tms function creates an Nx2 matrix (representing N artifacts) in cfg_ringing.artfctdef.tms.artifact or cfg_recharge.artfctdef.tms.artifact that contains the on- and offset of each marked/detected segment. Since we created two structures, we will combine both artifacts into one structure to do the rejection in one step. To achieve this we will create a new structure, cfg_artifact, that will contain artifact definitions for both the ringing/step response and the ringing artifact, each in separate subfields.
% Combine into one structure
cfg_artifact = [];
cfg_artifact.dataset = 'jimher_toolkit_demo_dataset_.eeg';
cfg_artifact.artfctdef.ringing.artifact = cfg_ringing.artfctdef.tms.artifact; % Add ringing/step response artifact definition
cfg_artifact.artfctdef.recharge.artifact = cfg_recharge.artfctdef.tms.artifact; % Add recharge artifact definition
The function ft_rejectartifact allows us to manipulate trials containing artifacts in several ways: First we can choose to reject the whole trial, but that is not appropriate here, since every trial contains TMS artifacts. Second, we can choose to replace the artifactual data with nans, something that may lead to problems using other functions that expect real numbers. Third - the approach we will take in this tutorial -, we can remove the artifacts by segmenting the trials into smaller pieces. We will exclude the parts of the data that contain the artifact and keep the other parts. We can later reconstruct the trial using the trial matrix (trl) we defined and saved earlier.
cfg_artifact.artfctdef.reject = 'partial'; % Can also be 'complete', or 'nan';
cfg_artifact.trl = trl; % We supply ft_rejectartifact with the original trial structure so it knows where to look for artifacts.
cfg_artifact.artfctdef.minaccepttim = 0.01; % This specifies the minimumm size of resulting trials. You have to set this, the default is too large for thre present data, resulting in small artifact-free segments being rejected as well.
cfg = ft_rejectartifact(cfg_artifact); % Reject trials partially
We have already read-in the data segments without the artifacts for you. You can download the data here. If you have downloaded the dataset, you can load it with:
load data_tms_segmented.mat
You can skip the following block of code if you have downloaded data_tms_segmented in the previous step. Else, you can read the artifact-free segments using
cfg.channel = {'all' '-5' '-mastoid L' '-mastoid R'};
cfg.reref = 'yes';
cfg.refchannel = {'all'};
cfg.implicitref = '5';
data_tms_segmented = ft_preprocessing(cfg);
We’ve now split-up our trials into segments free of ringing/step response, and recharging artifacts. You can see that this has increased the number of trials.
>> size(data_tms_segmented.trial)
ans =
1 900
Using ft_databrowser we can browse through both the segmented as well as the raw data. If we use the artifact definition we previously created we can easily browse to the segments we’ve marked as artifacts.
% Browse through the segmented data
cfg = [];
cfg.artfctdef = cfg_artifact.artfctdef; % Store previously obtained artifact definition
cfg.continuous = 'yes'; % represent the segments or trials as continuous data
ft_databrowser(cfg, data_tms_segmented);
Using the arrow-buttons beneath the ‘ringing’ and ‘recharge’ buttons we can browse to the marked artifacts.
% Browse through the raw data
cfg = [];
cfg.artfctdef = cfg_artifact.artfctdef;
cfg.dataset = 'jimher_toolkit_demo_dataset_.eeg';
ft_databrowser(cfg);
Independent Component Analysis
We now have our data segmented, removed ringing/step response and recharge artifacts. Our data still contains the exponential decay and the cranial muscle artifacts. We will attempt to attenuate these artifacts following an approach based on work by Korhonen et al. Removal of large muscle artifacts from transcranial magnetic stimulation-evoked EEG by independent component analysis. We will use a slightly adapted version of their manual artifact rejection approach. To this end we will decompose our data into independent components and reject components that capture artifacts we wish to attenuate, while taking care we do not remove non-artifactual data.
Running an ICA can take quite some time depending on the size of the data. Depending on your system processing this dataset will take about 15-30 minutes. Furthermore, running an ICA requires quite a lot of memory. In this case it requires an estimated 4GB of additional system memory. We therefore advise you to download the result comp_tms.mat. You can then skip the following segment of code.
As ICA is in fact a spatial filter, it relies on the artifacts having a stable topography in the data. If the topography changes during the experiment, your artifact may be captured in more than one, or two components and potentially cannot be captured in sufficient components at all. If you therefore know beforehand that the topography of your artifacts are different for parts of your data, you may have to apply the ICA separately for these parts. For example, if you are running an experiment where different locations are stimulated in different conditions, you could run the ICA separately for each location.
A stable TMS coil position increases the spatial stability of the artifacts, which is beneficial for estimating the artifact as independent component.
If you have downloaded comp_tms.mat, you can load the data with the following cod
load comp_tms.mat
You can skip the following block of code if you have loaded comp_tms in the previous step.
%% Perform ICA on segmented data
cfg = [];
cfg.demean = 'yes';
cfg.method = 'fastica'; % FieldTrip supports multiple ways to perform ICA, 'fastica' is one of them.
cfg.fastica.approach = 'symm'; % All components will be estimated simultaneously.
cfg.fastica.g = 'gauss';
comp_tms = ft_componentanalysis(cfg, data_tms_segmented);
save('comp_tms','comp_tms','-v7.3');
Memory issues
To capture the TMS artifacts in sufficient detail, the EEG is usually acquired at the highest sampling rate possible, which makes the datasets large. If you are having memory issues running the ICA you can downsample your data beforehand. Please be aware that prior to down sampling your data is filtered using a low pass FIR filter at roughly half the target sampling frequency (for example, if you downsample to 1000Hz, your data will be lowpass filtered at 500Hz). Use the following code to downsample your dat
% save the data in the original sampling frequency
save('data_tms_segmented','data_tms_segmented','-v7.3')
% resample into new data structure
cfg = [];
cfg.resamplefs = 1000; % Frequency to resample to
cfg.demean = 'yes';
data_tms_segmented_resampled = ft_resampledata(cfg, data_tms_segmented);
% remove the original data from memory
clear data_tms_segmented
Now you can run the ICA. After you have run the ICA on the downsampled data, reload the original data and apply the spatial unmixing matrix on the original data. The reason for this is that the data is demeaned prior to resampling to avoid artifacts at the edge of the trials. Since our trials are divided into segments, your trials may end up with strange baseline shifts because each segment is demeaned separately.
clear data_tms_resampled
load data_tms_segmented
After having run the ICA we are left with a structure similar to our data_tms_segmented structure with a few fields added and some changed.
>> data_tms_segmented
data_tms_segmented =
hdr: [1x1 struct]
label: {61x1 cell}
time: {1x900 cell}
trial: {1x900 cell}
fsample: 5000
sampleinfo: [900x2 double]
trialinfo: [900x1 double]
cfg: [1x1 struct]
>> comp_tms
comp_tms =
fsample: 5000
time: {1x900 cell}
trial: {1x900 cell}
topo: [61x60 double]
unmixing: [60x61 double]
label: {60x1 cell}
topolabel: {61x1 cell}
sampleinfo: [900x2 double]
trialinfo: [900x1 double]
cfg: [1x1 struct]
Instead of channels by time matrices, our data is now represented in component by time matrices, one for each trial (or segment in our case). The added fields .topo, .unmixing, and .topolabel contain the information necessary to back-project the components to the channel level.
We are now going to have a look at the timecourse of the components to identify the ones to be rejected. We will also look at a topographical representation of the components to see if they reflect spatial distributions indicating them to be an artifact (dipole, close to the site of stimulation). As we know that all our TMS-EEG artifacts are time-locked to the onset of the TMS-pulse, we can simplify the visual inspection by looking at the time-locked average representation of the components.
cfg = [];
comp_tms_avg = ft_timelockanalysis(cfg, comp_tms);
We can now browse the averaged data in the same way we browsed our channel data. The segments that we have excluded will be plotted as gaps. As with our channel data, you can either browse through the channels with ft_databrowser but also with MATLAB’s built-in plotting functions. Please be aware that if we are using ft_databrowser do browse averaged data (i.e. output from ft_timelockanalysis) we can only browse through channels as there are no trials.
figure;
cfg = [];
cfg.viewmode = 'butterfly';
ft_databrowser(cfg, comp_tms_avg);
Exercise: plotting components
Try to plot the components using MATLAB’s built-in plot function. Which do you prefer to browse the components?
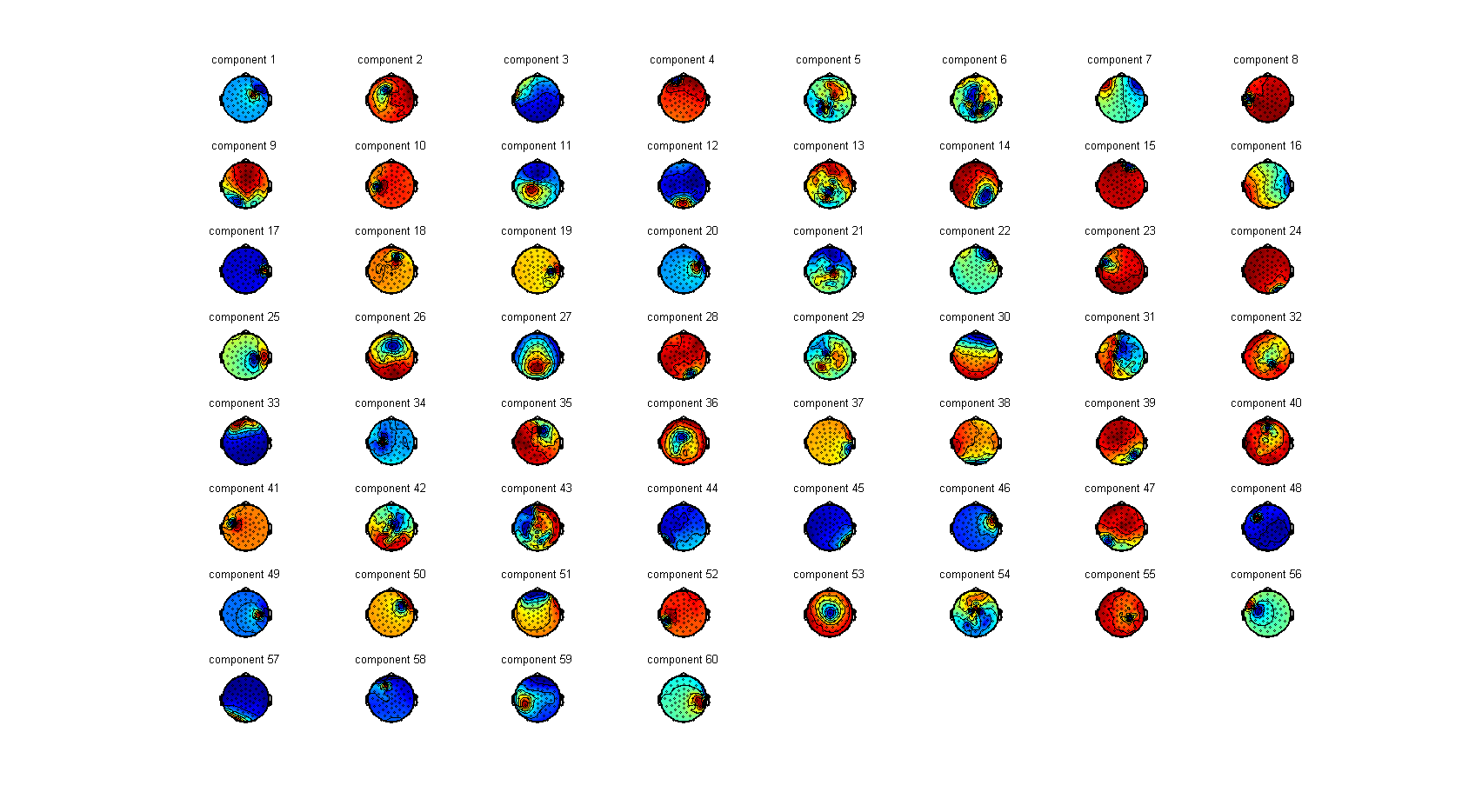
As ICA is in principle a spatial filter, we can inspect how each component loads spatially onto our original channel data. For this purpose we will use ft_topoplotIC.
figure;
cfg = [];
cfg.component = [1:60];
cfg.comment = 'no';
cfg.layout = 'easycapM10'; % If you use a function that requires plotting of topographical information you need to supply the function with the location of your channels
ft_topoplotIC(cfg, comp_tms);
Using ft_databrowser, or MATLAB’s plotting function, together with the output from ft_topoplotIC should be able to find one or two components that capture the decay artifact and/or the cranial muscle if the ICA was successful.
Exercise: find the components
Try to find components reflecting the decay and/or muscle artifact. Which ones would you remove?
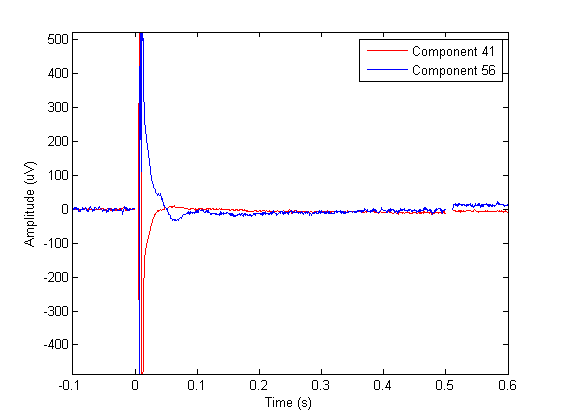
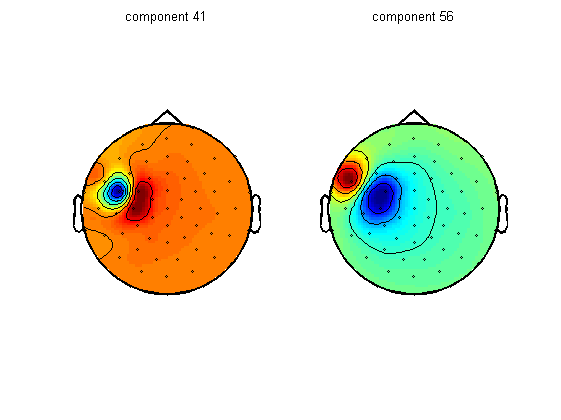
Due to various factors it is likely that you will not be able to fully capture both artifacts into one or two components. In this case, the decay artifact is captured pretty well by component numbers 41 and 56. The topographies of these components overlaps with the location of stimulation. These components also capture a large part of the cranial muscle artifact, but you can also see that almost every other component contains parts of cranial muscle artifacts. We can therefore conclude that the cranial muscle artifact cannot be fully removed by ICA in this dataset.
Exercise: removing other types of noise
At this stage you can also use your ICA data to remove other types of artifacts/noise. ICA is particularly well-suited to deal with eye-blinks and saccades, and can potentially remove other types of noise as well (also see the examples on EOG artifacts and ECG artifacts).
As these types of noise are not time-locked to onset of the TMS-pulse you can use ft_databrowser to browse through the trials in a component view. Be aware that in this case you are browsing the segments of the original trials.
cfg = [];
cfg.layout = 'easycapM10';
cfg.viewmode = 'component'; % Mode specifically suited to browse through ICA data
ft_databrowser(cfg, comp_tms);
Which components would you further suggest to remove?
We now have a list of components we want to remove:
- Decay & Muscle: 41, 56
- Saccades: 7
- Eye-blinks: 33
- Line noise: 31
- Maintenance recharging/Muscle: 1, 25, 52, 37, 49, 50
We can now remove these components from the data. Using ft_rejectcomponent you can remove components and transform the data back to a channel representation. First, however, we will need to revert a step we took before performing the ICA.
Keep in mind that our data is divided into segments of our original trials. Before performing ICA, the mean of each trial is subtracted from our data. This so-called demeaning is done to simplify the ICA algorithm. If we were to immediately back-transform our component data to a channel-representation and then restructure the segments into our original trials, we may have offsets in our trials because a different mean was subtracted for each segment. Especially the segment containing the decay artifact may be shifted a lot due to the large values in the period containing the decay.
Running an ICA results in a matrix that can be used to transform our original data into component data. We can now use this ‘unmixing matrix’ on our data without removing the mean as this was only necessary to produce the unmixing matrix. We will therefore apply this on our data without demeaning.
cfg = [];
cfg.demean = 'no'; % This has to be explicitly stated as the default is to demean.
cfg.unmixing = comp_tms.unmixing; % Supply the matrix necessay to 'unmix' the channel-series data into components
cfg.topolabel = comp_tms.topolabel; % Supply the original channel label information
comp_tms = ft_componentanalysis(cfg, data_tms_segmented);
Now we can remove the components.
cfg = [];
cfg.component = [ 41 56 7 33 1 25 52 37 49 50 31];
cfg.demean = 'no';
data_tms_clean_segmented = ft_rejectcomponent(cfg, comp_tms);
The components have now been removed and the data is back into its original channel representation
clear comp_tms % to free up system memory, clear the comp structure
We can now have a look at the current status of our data.
cfg = [];
cfg.vartrllength = 2;
cfg.preproc.demean = 'no'; % Demeaning is still applied on the segments of the trials, rather than the entire trial. To avoid offsets within trials, set this to 'no'
data_tms_clean_avg = ft_timelockanalysis(cfg, data_tms_clean_segmented);
% Plot all channels
for i=1:numel(data_tms_clean_avg.label) % Loop through all channels
figure;
plot(data_tms_clean_avg.time, data_tms_clean_avg.avg(i,:),'b'); % Plot all data
xlim([-0.1 0.6]); % Here we can specify the limits of what to plot on the x-axis
title(['Channel ' data_tms_clean_avg.label{i}]);
ylabel('Amplitude (uV)')
xlabel('Time (s)');
end
Exercise: how successful were we in removing TMS artifacts?
At the beginning we determined we had to deal with ringing/step response, cranial muscle, recharging and exponential decay artifacts. Have a look at the data, how successful were we in removing all of them? What is left and how could we deal with this?
Interpolation
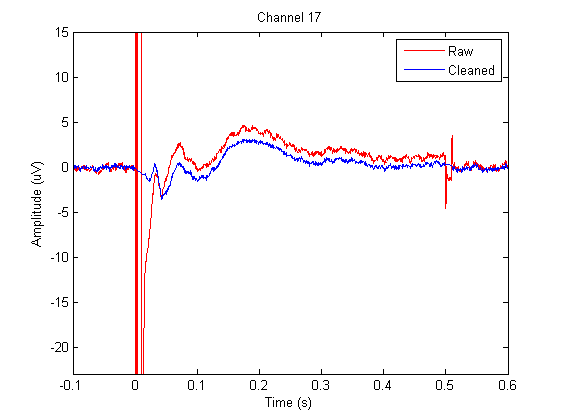
At this point we’ve removed the ringing/step response and discharge artifact by excluding it from our data. We’ve attenuated the exponential decay artifact using ICA. We’ve attenuated the muscle artifact a bit but have not succeeded as can be seen on the above plot of channel 17. Be aware that the ringing/step response artifact has been completely removed. The spiky part of the signal around 10ms, right after the gap, is purely a cranial muscle artifact. To prevent this artifact from interfering with our further processing steps we will also cut and interpolate the remainder of the muscle artifact.
Remember that our data is divided into segments of trials excluding the ringing/step response artifact and the recharging artifact. We are first going to recreate the original trial structure by combining the trial segments. The gaps that we’ve created by excluding certain segments will first be filled with nans. Afterwards all the segments containing nans will be interpolated. The period containing the muscle artifact will also be replaced by nans so that it can be interpolated as well.
We will first recreate our original trial structure. For this purpose we require the original trl matrix created at the beginning of the tutorial here.
% Apply original structure to segmented data, gaps will be filled with nans
cfg = [];
cfg.trl = trl;
data_tms_clean = ft_redefinetrial(cfg, data_tms_clean_segmented); % Restructure cleaned data
We’ve now recreated the original trial structure and put all pieces of data back into their original position. The gaps in the data due to the ringing and recharge artefacts have been willed with NaN, i.e. not-a-number values.
>> data_tms_clean
data_tms_clean =
hdr: [1x1 struct]
label: {61x1 cell}
fsample: 5000
trial: {1x300 cell}
time: {1x300 cell}
trialinfo: [300x1 double]
sampleinfo: [300x2 double]
cfg: [1x1 struct]
As you can see from the .trial field from both data structures we again have 300 trials. The cleaned dataset still contains the muscle artifact, we will therefore fill that segments with nans. The data (channel x time) for each trial is located in cells within the trial field in our data structure. We are now going to loop through each trial and replace the segment that contain the muscle artifact with nans. Browsing through the data it seems sufficient to replace up to 15ms after stimulation onset with nans.
When we want to know which data point corresponds to any given time point we often use the function nearest. This function finds the index number of a data point in a range of values that is closest to the point you request. For example, consider the vector [0 0.3 0.6 0.9 1.2]. If you would want to know the value closest to 0.5 the output nearest gives is 3, which stands for the third value in the vector: 0.6. This has advantages over MATLAB’s standard way of finding values within vectors (e.g., a==0.5) because we often do not exactly know the exact value we are interested in.
% Replacing muscle artifact with nans
% Specify the window we would like to replace with nans in seconds
muscle_window = [0.006 0.015];
% Find the indices in the time vector corresponding to our window of interest
muscle_window_idx = [nearest(data_tms_clean.time{1},muscle_window(1)) nearest(data_tms_clean.time{1},muscle_window(2))];
for i=1:numel(data_tms_clean.trial)
% Replace the segment of data corresponding to our window of interest with nans
data_tms_clean.trial{i}(:,muscle_window_idx(1):muscle_window_idx(2))=nan;
end
Now that everything we would like to interpolate has been replaced by nans we can start the interpolation. The function ft_interpolatenan loops through trials and channels and interpolates segments containing nans using MATLAB’s built-in interp1 function. You can therefore use all the methods supported by this function in your interpolation. We will use cubic interpolation as it avoids sharp transitions from your data to the edges of the interpolated segments you might create by using linear interpolation but does not appear to introduce strong artificial sinusoids as spline interpolation sometimes does. Feel free to try-out the different types of interpolation.
% Interpolate nans using cubic interpolation
cfg = [];
cfg.method = 'pchip'; % Here you can specify any method that is supported by interp1: 'nearest','linear','spline','pchip','cubic','v5cubic'
cfg.prewindow = 0.01; % Window prior to segment to use data points for interpolation
cfg.postwindow = 0.01; % Window after segment to use data points for interpolation
data_tms_clean = ft_interpolatenan(cfg, data_tms_clean); % Clean data
% compute the TEP on the cleaned data
cfg = [];
cfg.preproc.demean = 'yes';
cfg.preproc.baselinewindow = [-0.05 -0.001];
data_tms_clean_avg = ft_timelockanalysis(cfg, data_tms_clean);
We can now compare the raw data with the cleaned data. If you do not have the time-locked average of the raw data anymore, you can download it here and load it with:
load data_tms_avg
Using the following code we will compare the average TEP of the original data with the averaged TEP of the cleaned dat
for i=1:numel(data_tms_avg.label) % Loop through all channels
figure
plot(data_tms_avg.time, data_tms_avg.avg(i,:),'r'); % Plot all data
hold on
plot(data_tms_clean_avg.time, data_tms_clean_avg.avg(i,:),'b'); % Plot all data
xlim([-0.1 0.6]); % Here we can specify the limits of what to plot on the x-axis
ylim([-23 15]); % Here we can specify the limits of what to plot on the y-axis
title(['Channel ' data_tms_avg.label{i}])
ylabel('Amplitude (uV)')
xlabel('Time (s)')
legend({'Raw' 'Cleaned'})
end
After artifact removal, make sure to browse through your channels comparing the data prior and post correction to check if there are any residual artifacts left. It could be that you will have to interpolate a bit more or try to remove additional independent components.
At this point we’ve sufficiently cleaned our data of TMS artifacts so we can continue with the rest of our analysis. Only after this artifact removal is it safe to perform (post-)processing steps such as filtering, demeaning, detrending, and downsampling.
Post-processing
Now that we have cleaned the data we could apply some (post-)processing steps such as filtering, detrending, demeaning, and downsampling. At this point we are only going to downsample our data. Depending on your further analysis you may wish to apply other processing steps as well. It might be worth to note that some analysis steps may require different processing. For example, when looking at TMS evoked potentials (TEPs), you may want to filter your data to remove high-frequency noise. For performing time-frequency analysis this is not necessary but you would perhaps want to detrend your data, which is again not advised for analyzing TEPs. In short, different analysis methods may require different processing steps. Luckily the functions used to produce these analysis (e.g., ft_timelockanalysis and ft_freqanalysis) allow you to apply the same preprocessing steps to your input data as you can apply with ft_preprocessing. This allows you to apply separate processing steps suited for each analysis without having to create additional data structures.
For this reason we are only going to downsample our data and apply slightly different (post-) processing steps later on. Coincidentally, downsampling cannot be done with ft_preprocessing but with ft_resampledata.
cfg = [];
cfg.resamplefs = 1000;
cfg.detrend = 'no';
cfg.demean = 'yes'; % Prior to downsampling a lowpass filter is applied, demeaning avoids artifacts at the edges of your trial
data_tms_clean = ft_resampledata(cfg, data_tms_clean);
Now that we have cleaned our data and applied our (post-)processing steps we continue on with our analysis, but first we will save our data.
save('data_tms_clean','data_tms_clean','-v7.3')
When saving data we always use the switch ‘-v7.3’ as this allows files to be larger than 4GB, which is often the case in TMS-EEG data.
Analysis
Now that we have cleaned our data we can continue with our analyses. Initially we started out with the question whether pre-contraction affects the TMS-evoked potential. To address this question we are going to compare the amplitudes of the TEPs, inspect the frequency content of the response to TMS, and look at Global Mean Field Power.
If you do not have the output from the previous cleaning steps you can download the cleaned dataset here.
Time-locked averaging
Remember that we have two conditions to compare in the current dataset: ‘relax’ & ‘contract’. The condition was indicated in the dataset by markers placed in the EEG. When we read-in our data we based the trials on these markers, the information to which condition each trial belongs can therefore be found in the .trialinfo field of our data structure. In this field the ‘relax’ condition is indicated by the number 1 and the ‘contract’ condition by the number 3.
>> data_tms_clean.trialinfo
ans =
1
1
1
1
.
.
.
3
3
3
.
.
.
We can use the information in this field to perform timelock analysis on both conditions separately. Each row in the .trialinfo field corresponds to a trial in our dataset. If we therefore find all the rows corresponding to one number representing a condition, we know which trials belong to that condition.
In calculating the timelocked averages we will also apply a baseline correction (50ms to 1ms prior to TMS onset) and we will apply a lowpass filter of 35Hz.
cfg = [];
cfg.preproc.demean = 'yes';
cfg.preproc.baselinewindow = [-0.05 -0.001];
cfg.preproc.lpfilter = 'yes';
cfg.preproc.lpfreq = 35;
% Find all trials corresponding to the relax condition
cfg.trials = find(data_tms_clean.trialinfo==1);
relax_avg = ft_timelockanalysis(cfg, data_tms_clean);
% Find all trials corresponding to the contract condition
cfg.trials = find(data_tms_clean.trialinfo==3);
contract_avg = ft_timelockanalysis(cfg, data_tms_clean);
We are also going to calculate the difference between the two averages. To this end we will use the function ft_math, which allows you to perform a certain number of mathematical operations on one or multiple data structures.
% Calculate the difference ERP wave
cfg = [];
cfg.operation = 'subtract'; % Operation to apply
cfg.parameter = 'avg'; % The field in the data structure to which to apply the operation
difference_avg = ft_math(cfg, contract_avg, relax_avg);
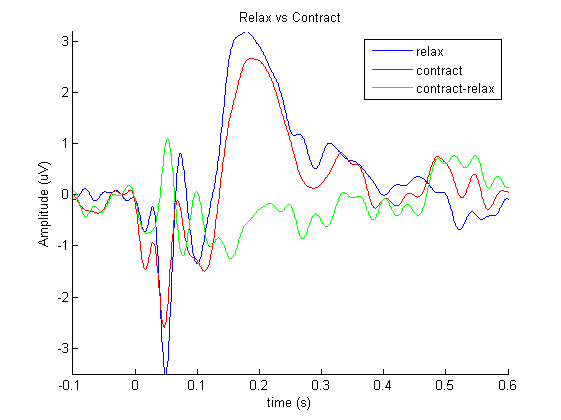
We will now plot both conditions and their difference using ft_singleplotER, a function ideally suited for plotting and comparing conditions.
% Plot TEPs of both conditions
cfg = [];
cfg.layout = 'easycapM10'; % Specifying this allows you to produce topographical plots of your data
cfg.channel = '17';
cfg.xlim = [-0.1 0.6];
ft_singleplotER(cfg, relax_avg, contract_avg, difference_avg);
ylabel('Amplitude (uV)');
xlabel('time (s)');
title('Relax vs Contract');
legend({'relax' 'contract' 'contract-relax'});
A nice feature of ft_singleplotER is that you can select a time range in your plotting window and click on it to produce a topographical representation of your amplitudes at that time point if you’ve specified a layout. You can also use the function ft_topoplotER for this.
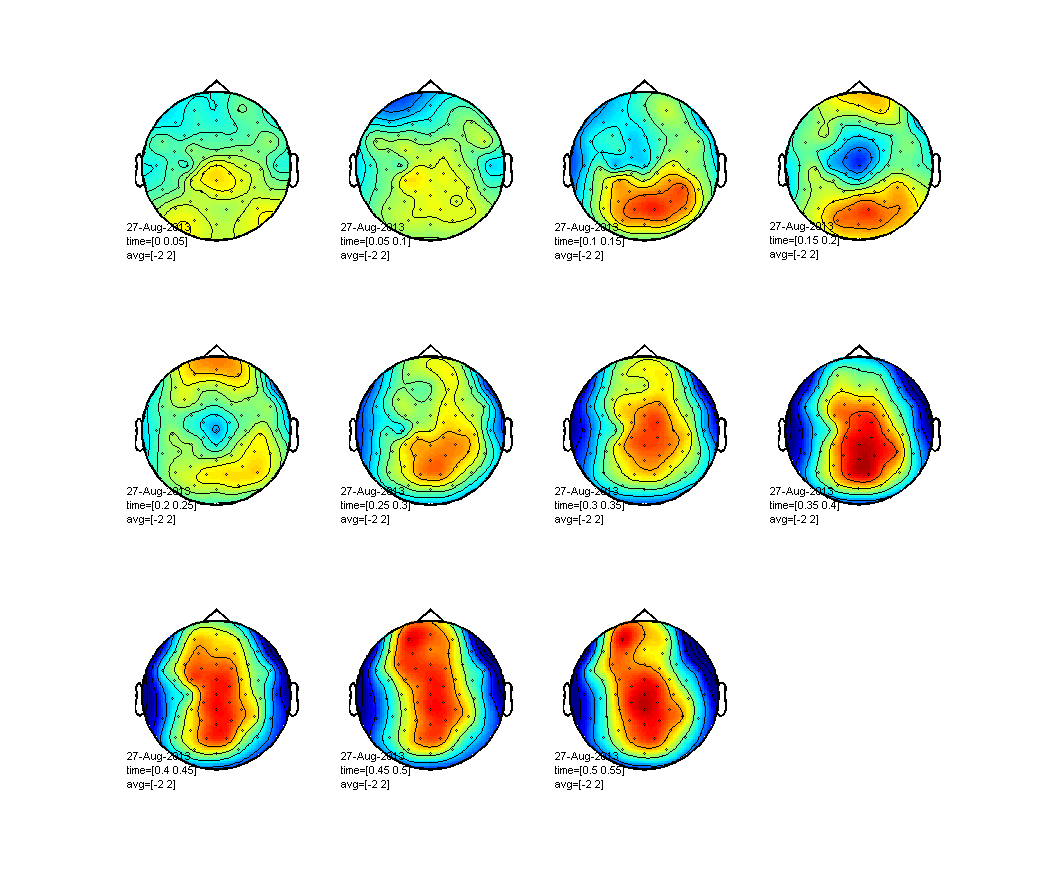
%% Plotting topographies
figure;
cfg = [];
cfg.layout = 'easycapM10';
cfg.xlim = 0:0.05:0.55; % Here we've specified a vector between 0 and 0.55 seconds in steps of 0.05 seconds. A topoplot will be created for each time point specified here.
cfg.zlim = [-2 2]; % Here you can specify the limit of the values corresponding to the colors. If you do not specify this the limits will be estimated automatically for each plot making it difficult to compare subsequent plots.
ft_topoplotER(cfg, difference_avg);
Exercise: where are the differences?
Where can you find the largest differences? How does the topography of these differences look like? Does it make sense given that we know we stimulated left-M1?
FieldTrip supports numerous ways of plotting your data. Each suited for a particular purpose. Most plotting functions can be subdivided into three categories, each category has a plotting function for a specific datatype. For us the following are the most useful
- ft_singleplotXXX for single channel data.
- ft_multiplotXXX plots data of all channels following the specified layout
- ft_topoplotXXX plots topographical distribution of a specified time-range over a 2D representation of the head
XXX can be ER, for event-related data or TFR for time-frequency data.
Please also have a look at Plotting data at the channel and source level for a tutorial on plotting data
Global Mean Field Power
Global Mean Field Power (GMFP) is a measure first introduced by Lehmann and Skandries (1979) and used by, for example, Esser et al. (2006) as a measure to characterize global EEG activity.
GMFP can be calculated using the following formula (from Esser et al. (2006))
where t is time, V is the voltage at channel i and K is the number of channels. In Esser et al. (2006) the GMFP is calculated on the average over all subjects. As we only have one subject, we will only calculate the GMFP within this subject. If you, however, have multiple subjects you can apply the same method but on the grand average (see for examples on handling multiple subjects: Parametric and non-parametric statistics on event-related fields). Basically, the GMFP is the standard deviation over channels.
FieldTrip includes the ft_globalmeanfield function to calculate the GMFP. This requires timelocked data as input. We will use similar preprocessing as applied in Esser et al. (2006).
% Create time-locked average
cfg = [];
cfg.preproc.demean = 'yes';
cfg.preproc.baselinewindow = [-0.1 -.001];
cfg.preproc.bpfilter = 'yes';
cfg.preproc.bpfreq = [5 100];
cfg.trials = find(data_tms_clean.trialinfo==1); % 'relax' trials
relax_avg = ft_timelockanalysis(cfg, data_tms_clean);
cfg.trials = find(data_tms_clean.trialinfo==3); % 'contract' trials
contract_avg = ft_timelockanalysis(cfg, data_tms_clean);
% GMFP calculation
cfg = [];
cfg.method = 'amplitude';
relax_gmfp = ft_globalmeanfield(cfg, relax_avg);
contract_gmfp = ft_globalmeanfield(cfg, contract_avg);
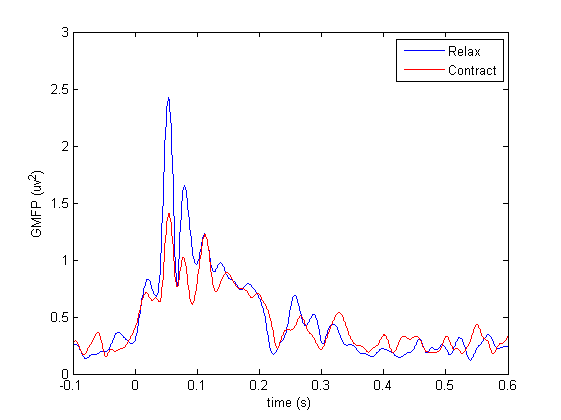
Now we can plot the GMFP of both conditions.
% Plot GMFP
figure
plot(relax_gmfp.time, relax_gmfp.avg,'b');
hold on
plot(contract_gmfp.time, contract_gmfp.avg,'r');
xlabel('time (s)')
ylabel('GMFP (uv^2)')
legend({'Relax' 'Contract'})
xlim([-0.1 0.6])
ylim([0 3])
Exercise: GMFP vs TEPs
Are there differences between the outcome of this analysis and the comparison between time-locked averages in the previous section? Can you see an advantage of using GMFP to compare conditions?
Time-frequency analysis
We have so far analyzed responses to the TMS pulse which always occur at the same time. Anything that is not phase-locked to the onset of the TMS pulse is cancelled out due to averaging. It is, however, possible that the TMS pulse induces responses that are not necessarily phase-locked to the onset of the pulse, for example changes in spontaneous oscillatory activity. To look at these induced responses we are going to look at time-frequency representations of our data. We will decompose our signals into frequencies and look at the averages of the power of these frequencies. Contrasting to time-lock analyses we are then sensitive to oscillatory activity not phase-locked to onset of the pulse (also see: Time-frequency analysis using Hanning window, multitapers and wavelets).
We will first decompose our signal into different frequencies using ft_freqanalysis. When doing spectral analyses it is important to detrend and demean your data prior to decomposing into frequencies to avoid strange looking powerspectra (see: Why does my TFR look strange (part I, demeaning)? and Why does my TFR look strange (part II, detrending)?). We will therefore detrend and demean our data using the .preproc option.
% Calculate Induced TFRs fpor both conditions
cfg = [];
cfg.polyremoval = 1; % Removes mean and linear trend
cfg.output = 'pow'; % Output the powerspectrum
cfg.method = 'mtmconvol';
cfg.taper = 'hanning';
cfg.foi = 1:50; % Our frequencies of interest. Now: 1 to 50, in steps of 1.
cfg.t_ftimwin = 0.3.*ones(1,numel(cfg.foi));
cfg.toi = -0.5:0.05:1.5;
% Calculate TFR for relax trials
cfg.trials = find(data_tms_clean.trialinfo==1);
relax_freq = ft_freqanalysis(cfg, data_tms_clean);
% Calculate TFR for contract trials
cfg.trials = find(data_tms_clean.trialinfo==3);
contract_freq = ft_freqanalysis(cfg, data_tms_clean);
We will also calculate the difference between conditions. Usually when plotting TFRs you can specify a baseline window. Since we are also calculating the difference between conditions and we are interested in the difference between both conditions AFTER baseline correction, we will first have to remove the baseline from our conditions.
% Remove baseline
cfg = [];
cfg.baselinetype = 'relchange'; % Calculate the change relative to the baseline ((data-baseline) / baseline). You can also use 'absolute', 'relative', or 'db'.
cfg.baseline = [-0.5 -0.3];
relax_freq_bc = ft_freqbaseline(cfg, relax_freq);
contract_freq_bc = ft_freqbaseline(cfg, contract_freq);
% Calculate the difference between both conditions
cfg = [];
cfg.operation = 'subtract';
cfg.parameter = 'powspctrm';
difference_freq = ft_math(cfg, contract_freq_bc, relax_freq_bc);
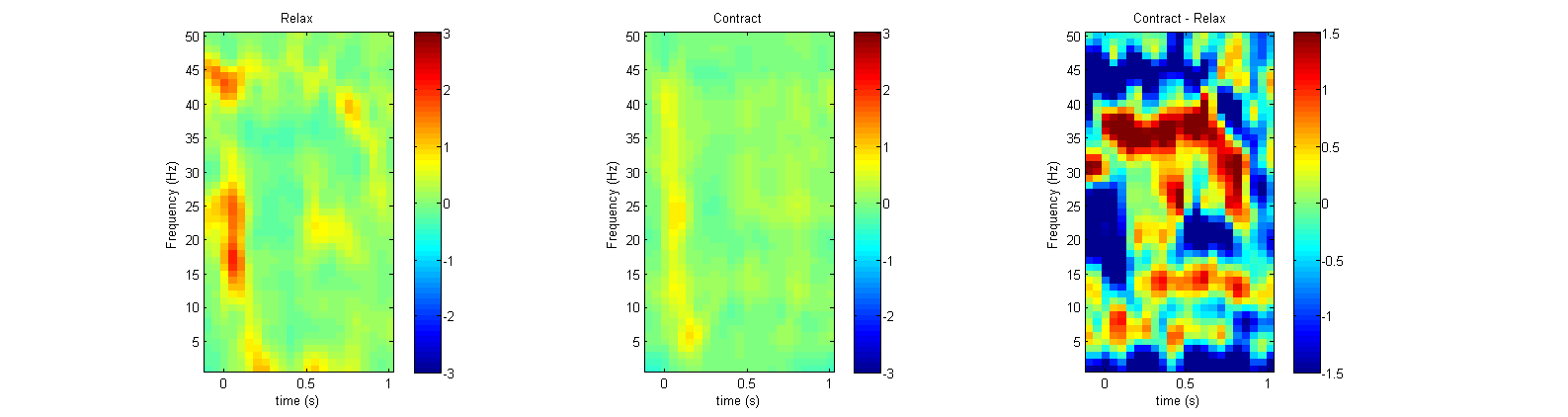
Now that we’ve calculated the TFRs for both conditions and their differences we can plot the results in various ways. We can start with plotting all TFRs on a 2D representation of the head using ft_multiplotTFR.
cfg = [];
cfg.xlim = [-0.1 1.0];
cfg.zlim = [-1.5 1.5];
cfg.layout = 'easycapM10';
figure;
ft_multiplotTFR(cfg, difference_freq);
This plot is fully interactive, click and drag to select one or more channels, click on them to view an averaged representation of the selected channels. You can also plot single (or multiple) channels in a single view using ft_singleplotTFR.
cfg = [];
cfg.channel = '17'; % Specify the channel to plot
cfg.xlim = [-0.1 1.0]; % Specify the time range to plot
cfg.zlim = [-3 3];
cfg.layout = 'easycapM10';
figure
% Use MATLAB's subplot function to divide plots into one figure
subplot(1,3,1)
ft_singleplotTFR(cfg, relax_freq_bc);
ylabel('Frequency (Hz)')
xlabel('time (s)')
title('Relax')
subplot(1,3,2)
ft_singleplotTFR(cfg, contract_freq_bc);
title('Contract')
ylabel('Frequency (Hz)')
xlabel('time (s)')
subplot(1,3,3)
cfg.zlim = [-1.5 1.5];
ft_singleplotTFR(cfg, difference_freq);
title('Contract - Relax')
ylabel('Frequency (Hz)')
xlabel('time (s)')
Exercise: additional information
What additional information can be gained by analyzing time-frequency data?
Exercise: conclusion
Now that we have described three ways of looking at our data, can we conclude the conditions differ? If so, how do they differ specifically?
Summary and suggested further reading
This tutorial covered how to deal with TMS artifacts in EEG in a single-pulse study. Furthermore, the tutorial showed three examples of how to further analyze this data with a certain research question in mind. In all examples two conditions were compared with each other. A next step would be to test whether differences between these conditions are statistically significant. To see how you can do this please have a look at the following tutorial